AlvaDesc v3.0.0 has been updated, it has a few new features but for Mac users it is now fully compatible with Apple Silicon!! AlvaDesc is
Tag: python
Sometimes when I’m comparing multiple datasets I end up with 10-20 different Vortex workspaces, for example if I’m comparing commercial screening collections from different vendors.
This looks interesting, handling large datasets is becoming more common and I’m always on the lookout for useful tools because pandas requires your dataframe to
As I’ve mentioned before the most popular page on this site is Fortran on a Mac, I’m not a big Fortran user but I do
This looks very useful for anyone having to process multiple molecules, I particularly like the error processing! The open-source package scikit-learn provides various machine learning
tmap is a very fast visualisation library for large, high-dimensional data sets. It was published in 2020 DOI and the code is available on GitHub
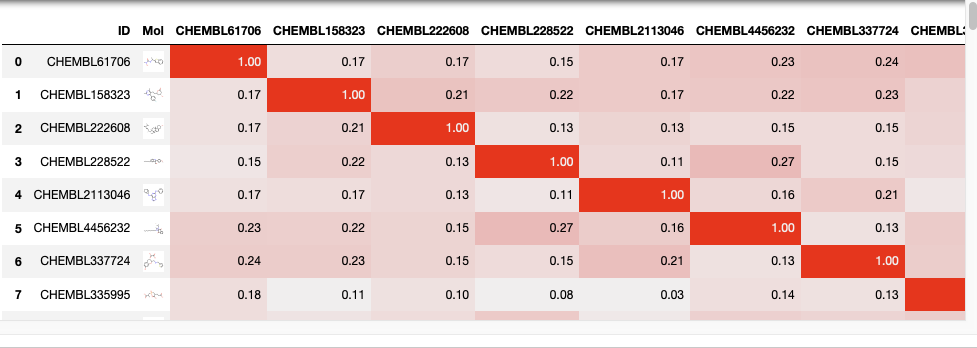
I’m sometimes asked for a tool to compare the similarity of a list of molecules with every other molecule in the list. I suspect there
Another tutorial from Greg Landrum, focussing on how to take advantage of modern multithreaded CPUs. https://greglandrum.github.io/rdkit-blog/posts/2024-02-11-more-multithreading.html Looking at Generating fingerprints, Molecular standardization, Conformer generation, RMSD
A previous post described Getting Ligand ID for multiple PDB files, using the PDBe API (https://www.ebi.ac.uk/pdbe/pdbe-services). The result is a csv file containing the ligand
The Protein Data Bank (https://www.rcsb.org) is an invaluable repository of 3D biomolecular structures. As of writing the database contains 214,791 structures (X-ray, Cryo-EM and NMR)