An interesting publication, using the IUPAC rules for naming compounds can be a challenge for for complex systems. This paper describes a transformer-based model https://doi.org/10.1186/s13321-024-00941-x
Category: Hints and Tutorials
Tutorials, Hints, Tips and Tricks
A reader asks about running VMD and installing Gromacs on M4 Macs. I don’t use either application and looking on the VMD website there is
Sometimes when I’m comparing multiple datasets I end up with 10-20 different Vortex workspaces, for example if I’m comparing commercial screening collections from different vendors.
The Jameel Clinic have just released Boltz-1, an open-source model designed to accurately model complex biomolecular interactions. Boltz-1 is an open-source model which predicts the
GitHub have announced that GitHub Copilot for Xcode is now available is now available for public preview. Key features of GitHub Copilot for Xcode: You

Fabulous blog post from Greg Landrum, Includes a tutorial on installing PostgreSQL and the cartridge with conda. This post shows how to use the RDKit
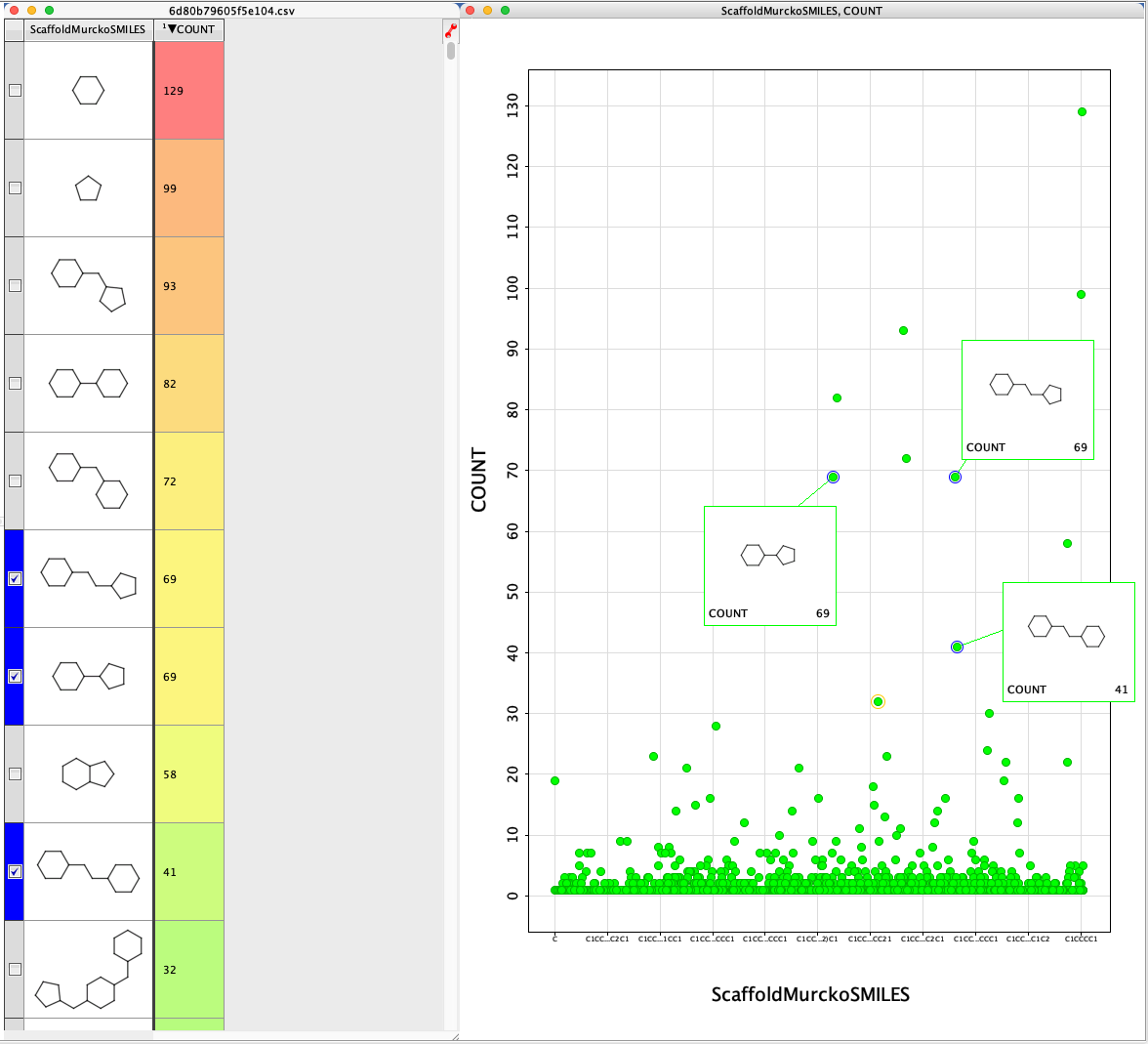
I published a page describing a Vortex script for category analysis that I wrote a while back but I thought I’d mention it because I’m
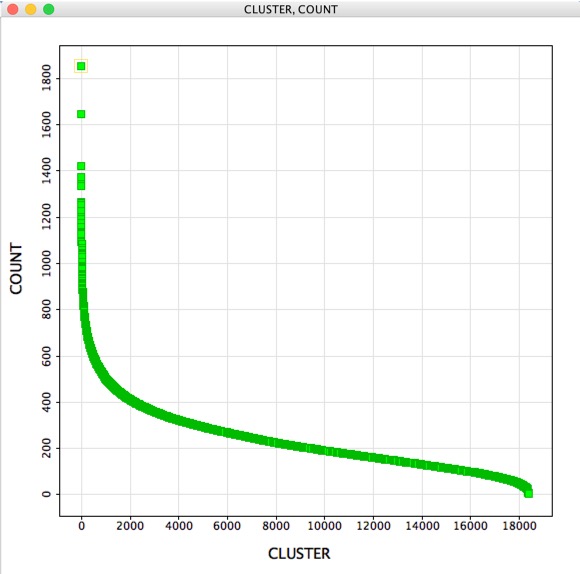
Clustering is an invaluable cheminformatics technique for subdividing a typically large compound collection into small groups of similar compounds. One of the advantages is that
The publication describing lwreg is now available. Here, we present lwreg, a lightweight, yet flexible chemical registration system supporting the capture of both two-dimensional molecular
An interesting alternative to hash-based folding on arXiv. https://arxiv.org/abs/2403.17954 Code is on GitHub https://github.com/MarkusFerdinandDablander/ECFP-substructure-pooling-Sort-and-Slice