You can download a beta version of Chemcraft here https://chemcraftprog.com/MacVersion.html Since this is not yet an Apple certified application read the installation instructions. Chemcraft is
Category: Macinchem Blog
The blog for the website
Generating conformations is always an issue, once there are multiple rotatable bonds then an exhaustive search becomes computationally intensive. So I always keep an eye
Mnova 15 is a major release that incorporates many new features in NMR, MSChrom, Mgears, ElViS, DB, new product versions such as Chrom Reaction Optimization 1.1, QC Profiling 1.2 and Multiplet Report 1.1, and the
Just stumbled across this blog on cheminformatics, machine learning (ML) and data science projects in drug discovery. Lots of useful code! Data in Life https://jhylin.github.io/Data_in_life_blog/
6 December 2023, 4-5.30pm UK,free/open to all HYBRID (@CCDC/on Zoom) Details & Registration: http://www.C-INF.net
A really interesting analysis of Apple silicon in particular the GPU design.
REINVENT is a molecular design tool for de novo design, scaffold hopping, R-group replacement, linker design, molecule optimization, and other small molecule design tasks. At
As virtual libraries get ever larger the challenges of virtual screening get larger. Whilst docking into a target protein is a very popular and successful
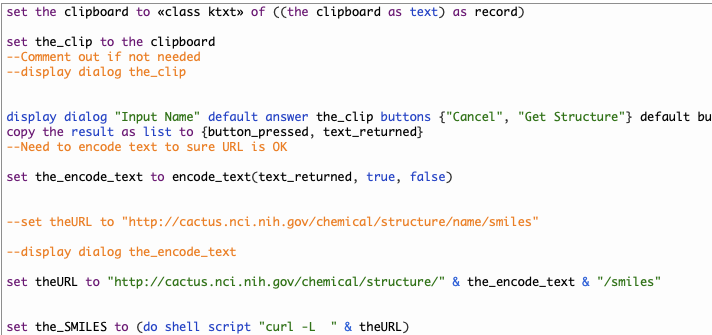
I’ve started adding a selection of useful applescripts, code can be copied from page or script downloaded. https://macinchem.org/category/applescript/ Applescript Description Print Clipboard This AppleScript prints

Programme Assessing Conformations of Small Molecules Against the Cambridge Structural Database Patrick McCabe, Cambridge Crystallographic Data Centre https://www.ccdc.cam.ac.uk/ SILVR: Guided Diffusion for Molecule Generation Nicholas