Rowan is a software platform that makes it possible to run high-level quantum chemical calculations through a web interface. In general, running quantum chemical calculations
Category: Jupyter Notebook
TabPFN is a foundation model trained on around 130,000,000 synthetically generated datasets that mimic “real world” tabular data. These datasets sampled dataset size and number
A very nice review of generative models for molecular design from Morgan Thomas. https://cheminformantics.blogspot.com/2024/12/structure-aware-generative-molecular.html Includes Jupyter notebooks for data and analysis.
I just stumbled across this and thought I’d flag it. VeloxChem is a Python-based open source quantum chemistry software developed for the calculation of molecular
A really interesting preprint caught my attention from Connor Coley’s group at MIT. ShEPhERD diffusing shape, electrostatics, and pharmacophores for bioisosteric drug design https://arxiv.org/abs/2411.04130v1 …
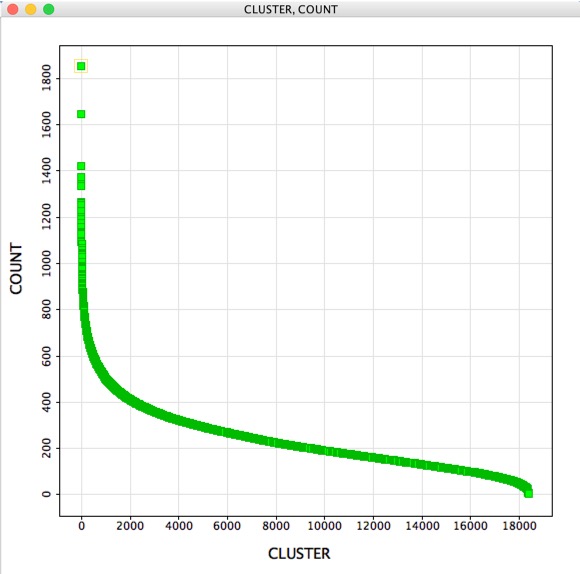
Clustering is an invaluable cheminformatics technique for subdividing a typically large compound collection into small groups of similar compounds. One of the advantages is that
tmap is a very fast visualisation library for large, high-dimensional data sets. It was published in 2020 DOI and the code is available on GitHub
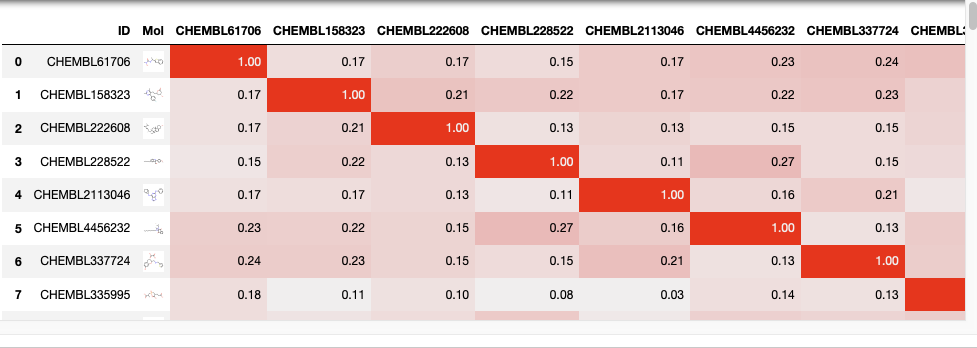
I’m sometimes asked for a tool to compare the similarity of a list of molecules with every other molecule in the list. I suspect there
Project Jupyter is the winner of the White House OSTP “Technical Advancement to Enable Open Science” category. Open science relies on technical advancements and infrastructures
Another tutorial from Greg Landrum, focussing on how to take advantage of modern multithreaded CPUs. https://greglandrum.github.io/rdkit-blog/posts/2024-02-11-more-multithreading.html Looking at Generating fingerprints, Molecular standardization, Conformer generation, RMSD