The Royal Society of Chemistry Chemical Information Group (CICAG) produce a twice-yearly newsletter highlighting news, features and reports about cheminformatics, computational chemistry, chemical information and
Author: chris
This morning I tried to print something to my HP LaserJet Pro printer and got an error. A quick search of the HP website suggested
New features in the Q-Chem 6.4 release include: Core-valence separation (CVS) scheme for ROCIS, XCIS, and QROCISfor calculating accurate core-level spectra of open-shell systems (Avik
Remote direct memory access (RDMA) is direct memory access from the memory of one computer into that of another without involving either computer’s operating system. This permits high-throughput, low-latency memory access over
An interesting meeting on Monday 16 March 2026 at Burlington House, London, UK The life sciences sector is undergoing a profound transformation. Advances inautomation, next-generation
Renaissance Philanthropy is releasing this call for AI for Science dataset proposals in collaboration with the UK’s Department for Science, Innovation and Technology (DSIT). This
One of the frequent situations after running a screen is you have a list of hits and you want to select related analogues to explore
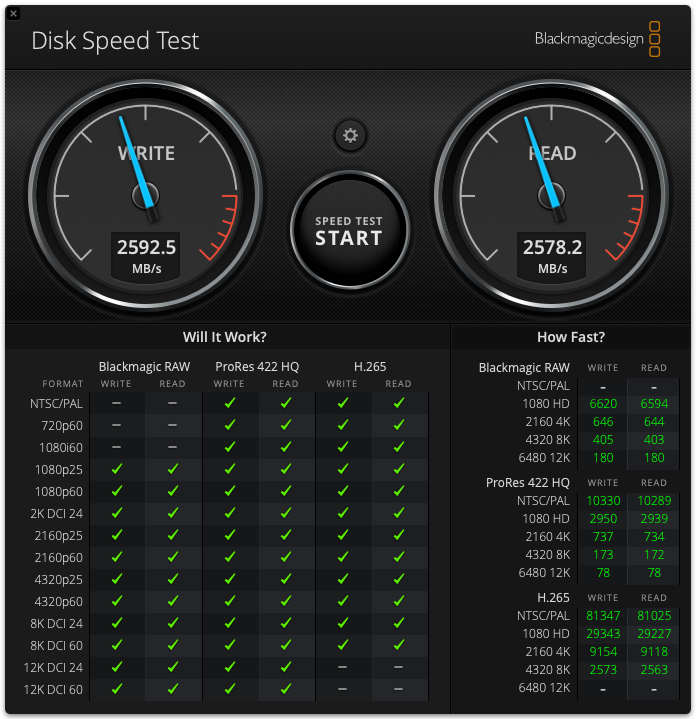
I’ve updated the page describing my testing of various external hard drive options. https://macinchem.org/wp-admin/post.php?post=1899 On my Mac Studio M2 Ultra the ACASIS 80Gbps M.2 NVMe
Next meeting 5-15 pm 4 December 2025, Lecture Theatre, Jeffrey Cheah Biomedical Centre, Puddicombe Way, Cambridge CB2 0AW. No need to register in advance –
This could be interesting… https://www.gov.uk/government/publications/ai-for-science-strategy/ai-for-science-strategy It is the execution that matters.