A Python Script
At the end of the previous page on setting up and accessing a Sqlite datasets from a Jupyter notebook there was and example of using a ChEMBLID to return the SMILES string. It is very easy to convert this to a python script as shown below.
After the imports, we first need to get the input, this is the first argument in the input command.
We then set up the connection to the base, note we need to include the full path to the database. Then construct the query and execute, parse the list returned to get the SMILES that the script returns
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 |
#!/usr/bin/env python # coding: utf-8 import sqlite3 import io import os import sys targetchembl = sys.argv[1] connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBLInChiKey/chembldata.db") cursor = connection.cursor() #targetchembl = "CHEMBL4741695" targetsmiles = cursor.execute("SELECT SMILES FROM ID_DATA WHERE CHEMBLID == ?", (targetchembl,),).fetchall() smilesreturn = targetsmiles[0][0] print(smilesreturn) |
Save this as ChEMBLIDLookup.py and you can then access it using the Terminal command shown below.
(base) chrisswain@ChrisM1MBP ~ % python /Users/chrisswain/Projects/ChEMBLInChiKey/ChEMBLIDLookup.py CHEMBL4741695
NC1=Nc2[nH]c(c(c2C(=O)N1)CN1CCN(CC1)Cc1cccc(c1)F)CN1CCN(CC1)Cc1cccc(c1)F
The real beauty though is this python script can be accessed from multiple source for example AppleScript
A ChemDraw Applescript to get structure
Open the Script Editor, it is probably in the Utilities folder within the Applications folder.
First create a dialog box and capture the response
|
1 |
set theResponse to display dialog "Get ChEMBL Structure" default answer "" with icon note buttons {"Cancel", "Search"} default button "Search" |
Then check which button was pressed and get the text returned (the ChEMBLID). Next construct the Terminal command, using the full path to the python that you want to use and the location of the python script. Copy the returned SMILES string to the clipboard.
If you are not sure of the path to python type “which python” in a Terminal window.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
set the button_pressed to the button returned of the theResponse if the button_pressed is "Search" then set theID to the text returned of the theResponse set thecommand to "/Users/chrisswain/miniconda3/bin/python /Users/chrisswain/Projects/ChEMBLInChiKey/ChEMBLIDLookup.py " & theID set theSMILES to do shell script thecommand --display dialog theSMILES set the clipboard to theSMILES else if the button_pressed is "Cancel" then end if |
Finally we bring ChemDraw to the front and and use the menu items scripting to paste as SMILES.
tell application “ChemDraw 22.0.0” activate
|
1 |
if enabled of menu item "Paste" then do menu item "SMILES" of menu "Paste Special" of menu "Edit" |
end tell
The full script is shown in the image below.
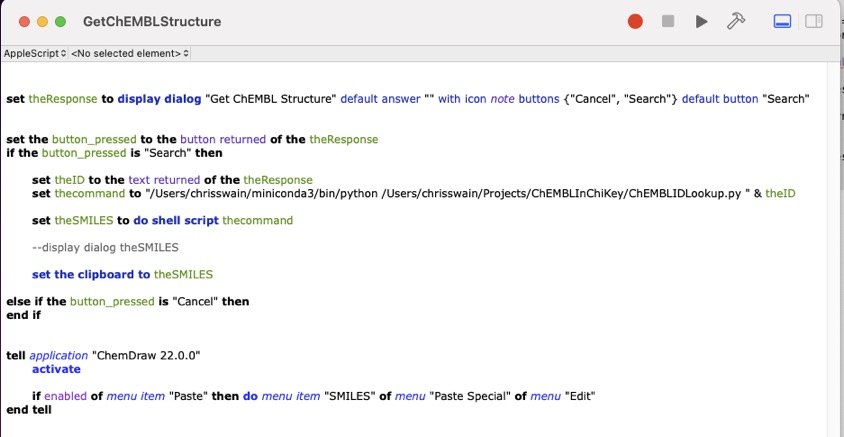
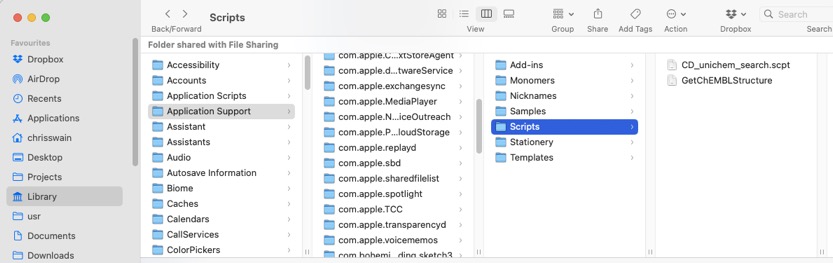
Save the script as GetChEMBLStructure in the ChemDraw scripts folder, the path to the folder is shown below (you might need to create it).
Next time you open ChemDraw you should see a Scripts dropdown menu in the top bar. Click on this and you should be able to choose the script.
This should open up a dialog box into which you can enter the ChEMBLID
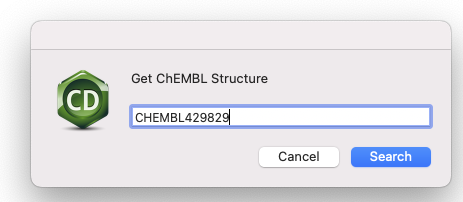
Click on the Search button and you should find the structure in the ChemDraw document
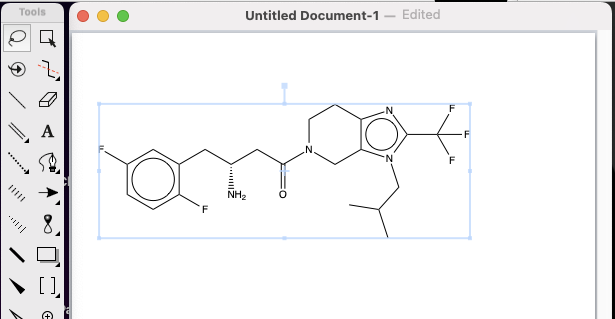
Checking if a structure is in ChEMBL
In a similar manner we can write a python script that takes a InChiKey as input and returns the CHEMBLID.
#!/usr/bin/env python
# coding: utf-8
import sqlite3
import io
import os
import sys
targetchembl = sys.argv[1]
connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBLInChiKey/chembldata.db")
cursor = connection.cursor()
#targetchembl = "CHEMBL4741695"
targetchemblID = cursor.execute("SELECT CHEMBLID FROM ID_DATA WHERE INCHIKEY == ?", (targetchembl,),).fetchall()
chemblreturn = targetchemblID[0][0]
print(chemblreturn)
Test the python script in the Terminal
(base) chrisswain@ChrisM1MBP ~ % python /Users/chrisswain/Projects/ChEMBLInChiKey/CheckChEMBLID.py GOLPEFAQLVNWBD-OEDNWNMQSA-N
CHEMBL414339
Now use ChemDraw to generate the InChiKey and pass it to the python script. The first part of the script checks if the Copy menu item is enabled to check a structure is selected. Then we script the menu items to copy the INChiKey to the clipboard. As before we create the SQLite command and capture the response. We then run the command and check the response, if the ChEMBLID is found it is displayed if not there is an error and the dialog displays “Not Found”. Finally the ChEMBLID is copied to the clipboard so you can paste it into the ChemDraw document.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 |
--set theInchikey to "" tell application "ChemDraw 22.0.0" activate if not (enabled of menu item "Copy") then do menu item "Select All" of menu "Edit" do menu item "InChI Key" of menu "Copy As" of menu "Edit" set theInchikey to the clipboard else do menu item "InChI Key" of menu "Copy As" of menu "Edit" set theInchikey to the clipboard end if end tell set thecommand to "/Users/chrisswain/miniconda3/bin/python /Users/chrisswain/Projects/ChEMBLInChiKey/CheckChEMBLID.py " & theInchikey try set theChEMBLID to do shell script thecommand display dialog theChEMBLID on error display dialog "Not Found" set theChEMBLID to "Not Found" end try set the clipboard to theChEMBLID |
Save it in the Chemdraw scripts folder and when you start up chemdraw it should be visible in the dropdown menu. Draw a structure and select the script and you should see a dialog
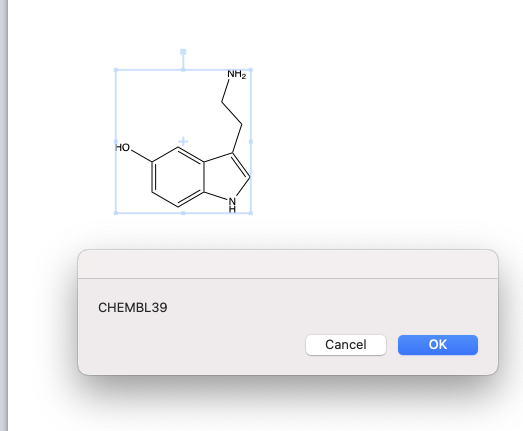
Since the ChEMBL ID is copied to the clipboard you can paste it under the structure.
Last updated 12 April 2023
2 thoughts on “Other ways of using the exact search”
Comments are closed.