One of the frequent situations after running a screen is you have a list of hits and you want to select related analogues to explore structure-activity around the screening hit, this is often called hit expansion. Whilst you can take each hit in turn and run a similarity search to identify potential follow up compounds from commercial sources, it would be nice to be able to do this a little more automatically. This Vortex script does this, many thanks to Dan Ormsby for his invaluable help.
So if you have a list of screening hits you would like to expand, import them into a Vortex workspace. Then import the collection of molecules you would like to select from into a new workspace, typically this might be a vendor catalogue.
Now run the script, a dialogue appears asking the user to choose the workspaces to compare (there needs to be a unique identifier for each molecule, I used a field called ID). Choose the two workspaces and set the similarity threshold.
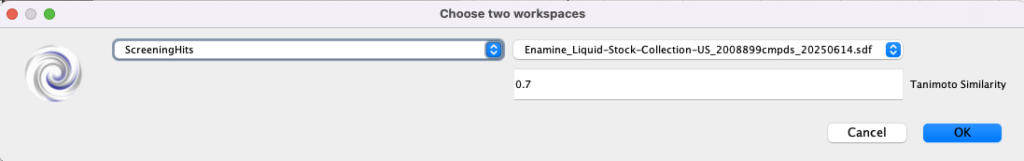
The results are shown in the image below, the original screening hit is SMILES2 and the suggested molecules for hit expansion are SMILES1. On my machine, expanding 10 hits selecting from 2M Enamine compounds takes little more than a screen refresh.

If you want to see how many similar molecules have been identified for each screening hit you can use the generic category script https://macinchem.org/2023/03/11/a-collection-of-vortex-scripts-to-aid-cluster-analysis/

You can download the script here