A previous post described Getting Ligand ID for multiple PDB files, using the PDBe API (https://www.ebi.ac.uk/pdbe/pdbe-services). The result is a csv file containing the ligand PDB id, that can be used to download the molecular structures from the PDB as shown below in a Jupyter Notebook.
The first cell simply imports the needed libraries.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
# Use PDB ID to download PDB Ligands # Authored by Chris Swain (http://www.macinchem.org) # Copyright CC-BY import csv import os import sys # Python 2 and 3 compatibility if sys.version_info[0] == 2: from urllib import urlretrieve else: from urllib.request import urlretrieve |
The next step is to identify the file containing the PDBid and import them (you will need to edit the path to the file). If you are using the file generated using the previous jupyter notebook it should be in the correct format. We also define folder into which the ligands will be downloaded.
|
1 2 3 4 |
# You may want to edit these parameters # File containing comma-separated list of the desired PDB IDs pdb_codes_file = '/Users/username/Desktop/PDBids_Ligands.csv' |
The input file format is shown below.
|
1 2 3 4 5 |
#Input file format, first element is protein ID, others are ligands #1L9K,SAH #1R6A,RVP,SAH #2JHP,GUN,SAH #2OY0,SAH |
The final cell is where the files are actually downloaded, first we read in the data from the input file, then looping through the file line by line. The first element on each line is the protein PDBid, we use this to create a folder within the downloads named as the protein PDBid.
|
1 |
download_folder = "PDBLigands/" + x[0] |
We then loop through the rest of the line starting at the second element to get the Ligand PDBid, use it to construct URL and download the file. All files are downloaded as compressed zip files.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 |
# Read the PDB IDs from the input file with open(pdb_codes_file) as f: pdb_records = f.read().split('\n') for i in pdb_records: x = i.rsplit(',') download_folder = "PDBLigands/" + x[0] try: os.makedirs(download_folder) except OSError as e: # Ignore OSError raised if it already exists pass for pdbid in x[1:]: filename = '%s_ideal.sdf' % pdbid[:4] # Add .gz extension if compressed url = 'https://download.rcsb.org/batch/ccd/%s' % filename destination_file = os.path.join(download_folder, filename +".zip") # Download the files try: urlretrieve(url, destination_file) except Exception as e: pass |
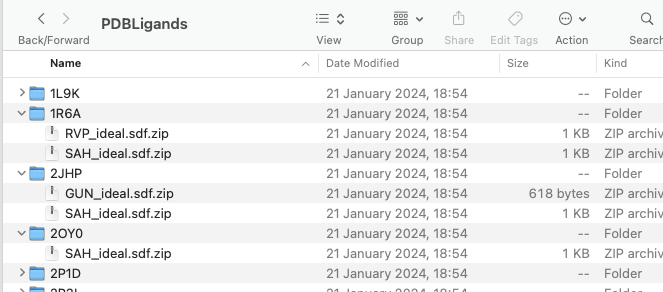
The result should be a folder called PDBLigands containing sub-folders for each of the individual protein PDBid and with each sub-folder the ligands have been downloaded.

You can download the Jupyter Notebook here.