TabPFN is a foundation model trained on around 130,000,000 synthetically generated datasets that mimic “real world” tabular data. These datasets sampled dataset size and number
Tag: rdkit
A really interesting preprint caught my attention from Connor Coley’s group at MIT. ShEPhERD diffusing shape, electrostatics, and pharmacophores for bioisosteric drug design https://arxiv.org/abs/2411.04130v1 …

Fabulous blog post from Greg Landrum, Includes a tutorial on installing PostgreSQL and the cartridge with conda. This post shows how to use the RDKit

The 2024.09.1 version of the RDKit was released on the 27th of September and is available via condo-forge (https://anaconda.org/conda-forge/rdkit). Some info on what is new
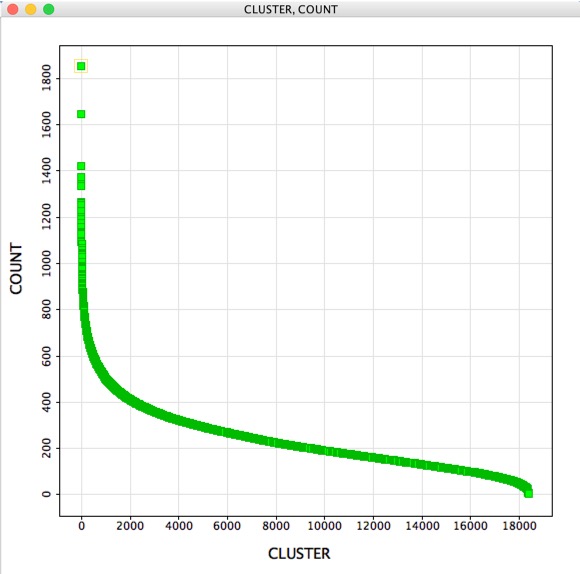
Clustering is an invaluable cheminformatics technique for subdividing a typically large compound collection into small groups of similar compounds. One of the advantages is that
The publication describing lwreg is now available. Here, we present lwreg, a lightweight, yet flexible chemical registration system supporting the capture of both two-dimensional molecular
I first mentioned this utility back in March and I’ve been using it since then and I have to say I find it invaluable. Clipboard-to-SMILES-Converter
rdEditor is a simple RDKit molecule editor GUI using PySide2. Code is on GitHub https://github.com/EBjerrum/rdeditor?tab=readme-ov-file The paper is on chemrxiv https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c
This looks very useful for anyone having to process multiple molecules, I particularly like the error processing! The open-source package scikit-learn provides various machine learning
tmap is a very fast visualisation library for large, high-dimensional data sets. It was published in 2020 DOI and the code is available on GitHub