Prediction of the metabolism of small molecules is very challenging and so having a variety of different tools is always useful. I’ve previously written Vortex
Category: Vortex Scripts
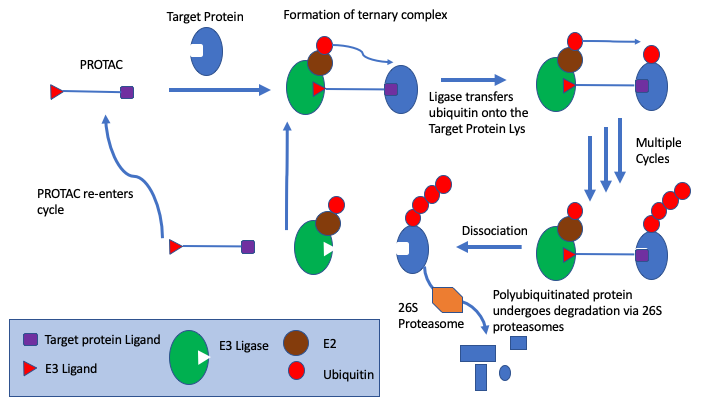
PROteolysis TArgeting Chimeras (PROTACs) technology provides an alternative to module biological function by specially using the ubiquitin proteasome system to induce degradation of the target
At least on a Mac the default place to store Vortex scripts is in the vortex folder ~/vortex/scripts Any folder/scripts you put in the vortex/scripts
One of the frequent situations after running a screen is you have a list of hits and you want to select related analogues to explore
When making selections from large datasets it is worth mentioning that as datasets get larger a simple random selection is often the best (and quickest)
Sometimes when you import a dataset into a Vortex workspace the default display can be not ideal. For example I imported this CDK7 dataset from
JDBC, which stands for Java Database Connectivity, is a Java API that allows Java applications to interact with relational databases. There are JDBC drivers for most
The previous scripts have allowed the user to get more information about a PDB entry and to import the ligand structures. This script allows the
The two previous vortex scripts used the PBD id to download information about a biomolecular structure structure from the Worldwide Protein Data Bank. The second
The Worldwide Protein Data Bank collects, organises and disseminates data on biological macromolecular structures. The wwPDB Partners are: PDBe, RCSB PDB, PDBj, BMRB, EMDB. Currently the PDB contains over