At least on a Mac the default place to store Vortex scripts is in the vortex folder ~/vortex/scripts Any folder/scripts you put in the vortex/scripts
Blog
Interesting post from ChEMBL team. We are excited to announce the start of the LIGAND-AI project, a 5-year project involving 18 partners to find ligands
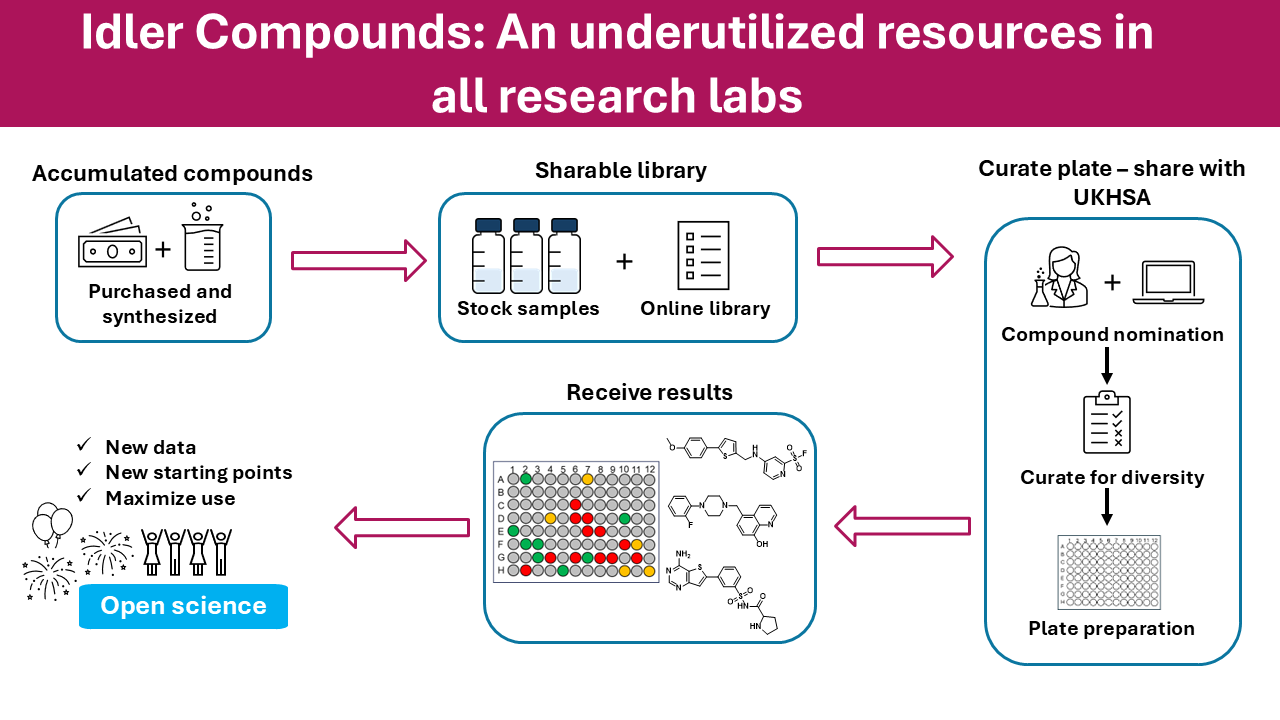
There was an interesting publication from the Todd group at UCL on Chemrxiv “Idler Compounds: A Simple Protocol for Openly Sharing Fridge Contents for Cross-Screening”

The life sciences sector is undergoing a profound transformation. Advances in automation, next-generation analytical technologies, and computational chemistry are redefining how research is conducted, making
LigandExtractor can be used to find all ligands in a file from the PDB. In addition to finding all possible ligands, it annotates any problems
Next up Date: 5 February 2026 Lecture Theatre, Jeffrey Cheah Biomedical Centre, Puddicombe Way, Cambridge CB2 0AW. No need to register in advance – just
The Royal Society of Chemistry Chemical Information Group (CICAG) produce a twice-yearly newsletter highlighting news, features and reports about cheminformatics, computational chemistry, chemical information and
This morning I tried to print something to my HP LaserJet Pro printer and got an error. A quick search of the HP website suggested
New features in the Q-Chem 6.4 release include: Core-valence separation (CVS) scheme for ROCIS, XCIS, and QROCISfor calculating accurate core-level spectra of open-shell systems (Avik
Remote direct memory access (RDMA) is direct memory access from the memory of one computer into that of another without involving either computer’s operating system. This permits high-throughput, low-latency memory access over