When AlphaFold was originally published by Google/Deepmind it was a step change in predicting protein 3D structures and it sparked an upsurge in activity around protein structure prediction leading to AlphaFold2 and including the collaboration with the EBI to produce the AlphaFold Protein Structure Database containing over 200 million entries, covering the human proteome and for the proteomes of 47 other key organisms important in research and global health.
Source code was made available on GitHub https://github.com/google-deepmind/alphafold
The latest update AlphaFold 3 claims to bring number of enhancements, including far greater accuracy on protein-ligand interactions, much higher accuracy on protein-nucleic acid interactions, and significantly higher antibody-antigen prediction accuracy. It seems that the major change is AlphaFold 3 assembles its predictions using a diffusion network, similar to those found in AI image generators. This is the result of work between Google/Deepmind/Isomorphic Labs, and this seems to have lead to a change in the publishing policy. Source code is no longer available, and access via a website (https://golgi.sandbox.google.com/about) is limited to non-commercial research only.
Publishing without source code makes it impossible for other scientists to check the work, and it does seem some reviewers were not happy with this https://twitter.com/RolandDunbrack/status/1788239357536858134
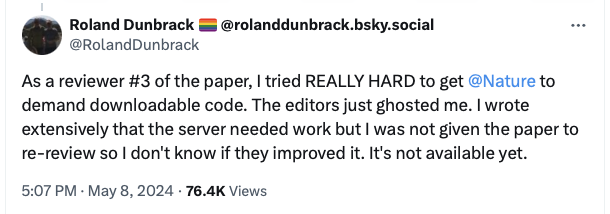
However, I suspect Isomorphic Labs see this as a strategic advantage for them and so want to keep it in house or collaborate with other pharma companies.
It is also worth noting there are a number of open-source protein structure prediction tools
OpenFold (https://openfold.io)
RoseTTAFold (https://github.com/RosettaCommons/RoseTTAFold)
Uni-Fold (https://github.com/dptech-corp/Uni-Fold)
OmegaFold (https://github.com/HeliXonProtein/OmegaFold)
EquiFold (https://github.com/Genentech/equifold).
rgn2 (https://github.com/aqlaboratory/rgn2)
IgFold (https://github.com/Graylab/IgFold).
