I’ve been interacting with Molecular Forecaster https://molecularforecaster.com and they have a number of very cool technologies to provide scientists with desktop tools for drug discovery. One of the really appealing features is that it is built in C++ and has a front-end JavaScript-based GUI. The code can be executed from the command-line and scripted to integrate into any workflow. They use standard input and output file formats to make integration quick and easy.
One of the tools that caught my eye was IMPACTS (In-silico Metabolism Prediction by Activated Cytochromes and Transition States) is a hybrid site-of-metabolism (SoM) identification tool which combines docking to CYP enzymes, transition state (TS) modeling, and rule-based substrate reactivity prediction to predict the SoM of xenobiotics. So I asked if we could to go through the analysis of the metabolism of Emend (Aprepitant) as I done for a couple of other tools
FAME2 https://www.macinchem.org/reviews/fame/fame2.php
CypReact https://www.macinchem.org/reviews/cypreact/cypreact.php
IMPACTS runs on macOS (64-bit only) and requires Java 1.6 and Xterm https://www.xquartz.org. Across 4 of the most import CYP isoforms (1A2, 2D6, 2C9, 3A4) in drug discovery, IMPACTS has showed excellent accuracies, ranging from 75% (considering the top2 metric) for CYP3A4 to 82% for 2C9. Overall, on average, the accuracy of IMPACTS is 77%. Importantly, the tool was tested on over 700 drugs and drug-like compounds for broad chemical space coverage. While IMPACTS has been internally tested on 4 major isoforms, protein files for 7 CYP isoforms (1A2, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4) are available.
he input is a 3D structure with the correct protonation state, Molecular Forecaster have their own drawing tools but since the file format is .mol2 these could be generated using a variety of alternative tools
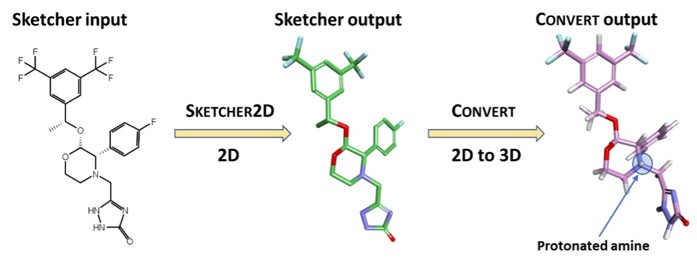
Once the molecule is in 3D mol2 format, it can be prepared for docking using SMART. SMART assigns to each atom a host of information, including a specific atom type, atomic charge, hybridization, and activation energy based on pre-computed values at the B3LYP/6-31G* level of theory. This information will be used by IMPACTS when modeling the transition state of the metabolic reaction. In this example, we will choose CYP3A4, which is the known major metabolizer of Aprepitant, along with CYP2C9.
When the IMPACTS module is reached, an “output” folder will automatically is created. The files created by IMPACTS will be stored here, and they include: a file tracking the progress of the calculation (.out), a file tracking errors and warnings (.log), a file summarizing the results (-results.txt), a file containing the TS of each predicted metabolite (.sdf format but in 3D), and a second metabolite file containing the metabolite after reaction. The results are shown below.
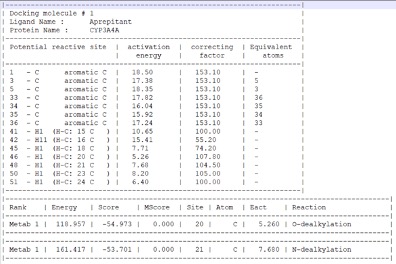
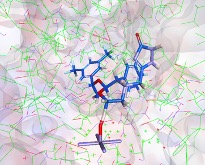
I really like the transition state output, this can viewed by any 3D molecule viewer. You can view the molecule itself or within the CYP active site.
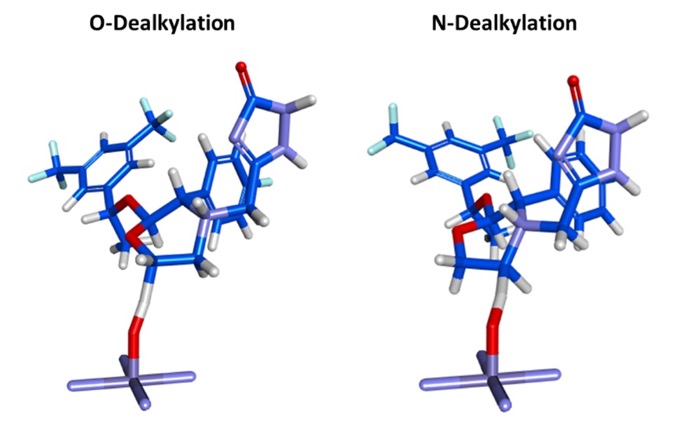
This allows the scientists the opportunity to look for possible modifications that might modulate the activity.
Summary
IMPACTS is a powerful state of the art tool for predicting sites of metabolism. The combination of docking to CYP enzymes, transition state (TS) modeling, and rule-based substrate reactivity prediction offers greater insight than many of the alternative tools. Of course this is only looking at CYP-mediated metabolism and there are many other non-CYP potential metabolic enzymes.
Worth reading
Campagna-Slater V., Pottel J., Therrien E., Cantin L.-D., Moitessier N., J Chem Inf Model, 2012, 52, 2471-2483 DOI
Last Updated 7 December 2022

