I was at the Dotmatics UGM recently and they gave an insight into some of the future directions. One of the areas under consideration is the use of Vortex support for Biological data analysis. This is only available in the latest version of Vortex, and must be first activated in the Vortex preferences.
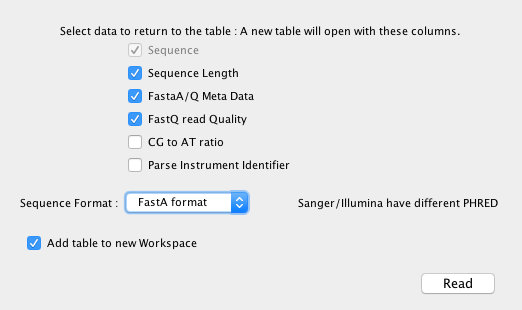
Once activated, “Open Sequence” is available from the “File” menu, this opens a dialog that allows you to define the import format (supports text, FastA, FASTQ Sanger, FASTQ Illumina).
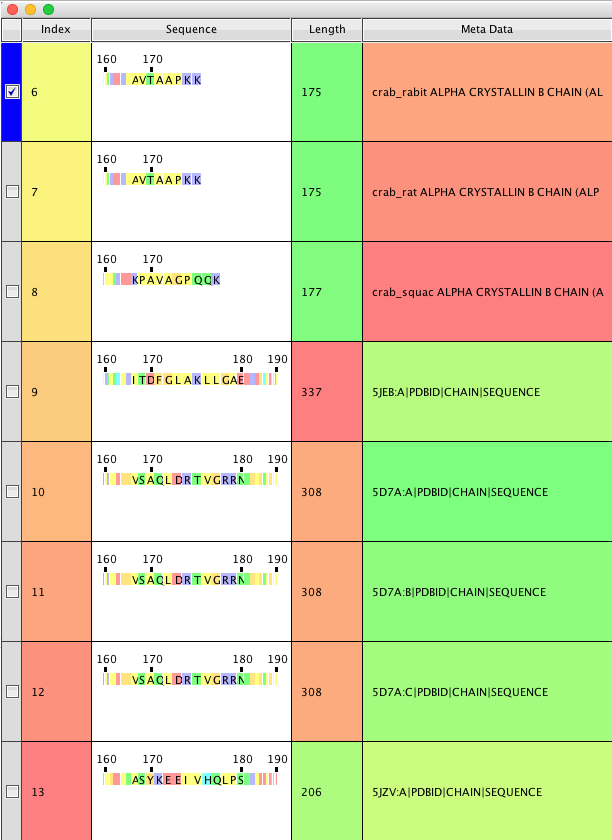
A table is created that contains an index column (ie to order with) and the selected return items;
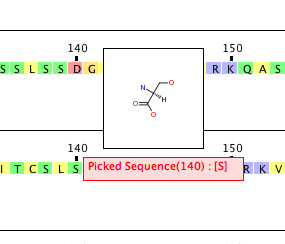
Clicking on one of the residues pops up a window showing the structure whilst clicking and dragging expands the selected portion of the sequence to allow focus on a specific region.
Currently there are a couple of tools provided, available from the “Tools” menu.
- Sequence Alignment
- Sequence Clustering
- Prosite Search
- Sequence Cross-Align
Selecting “Sequence Alignment” opens the dialog below, the tool automatically checks the biopolymer type and gives the appropriate options. You can either paste in a sequence or choose the first sequence in the table. The first option allows you to ignore the character case of the alignment, the default is to honour the case so that A ≠ a; so can be used to mask sections of sequence if necessary.
There are 4 alignment algorithms, the default is a fast global search (which is still a local alignment algorithm) which will return alignments between sections of sequence that show alignment and tries to optimise the output to cover the whole sequences. The fast local alignment will return multiple alignment hits for sections of aligned sequence optimised over local regions; the aligned results show the positional offsets of the alignments. The two slow methods force alignment throughout the sequence even if they don’t really align. All methods use blosum alignment matrices unless “Exact” is selected, then alignment is only matched were residues match .
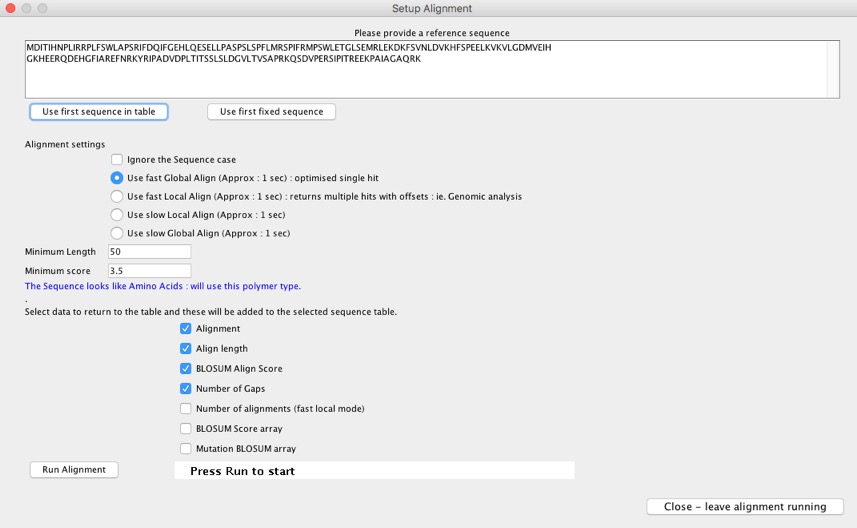
The results are added to the table as a series on added columns, Alignment a column where the alignment is shown, Align length the length of the alignment, Align Score , Number of gaps how many gaps within the aligned region. There is an option to run the alignment in the background but in my tests it was almost instantaneous. As before you can click on residues or sweep through the aligned sequences. Clicking on a residue displays the structure, it might be nice if clicking on a residue in one sequence also displayed the corresponding residue in the other sequence, useful in cases where residues are not exact matches.
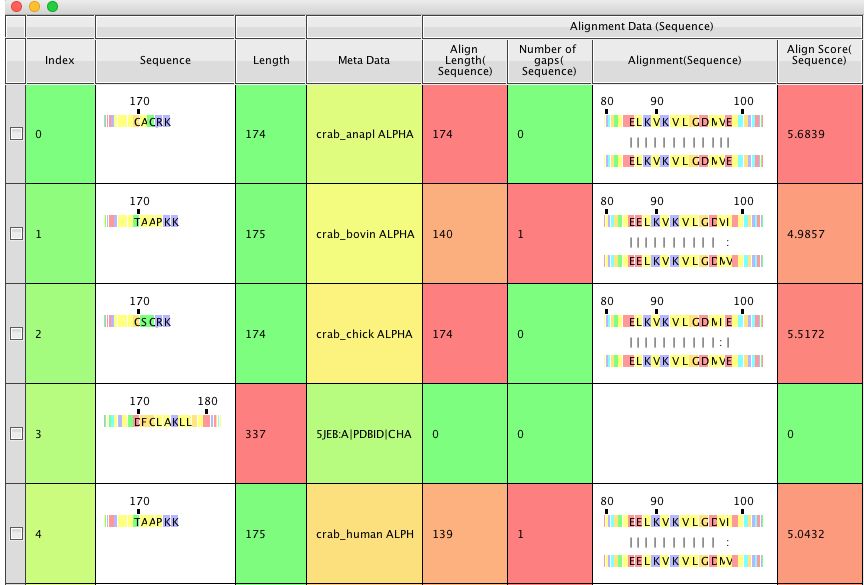
If found that you can rename the Align length, Number of Gaps and Align Score columns but not the Alignment Sequence. This means if you can run a second alignment and new columns for the Align length, Align Score, and Number of gaps will be generated, however the Alignment Sequence will only display the last alignment.
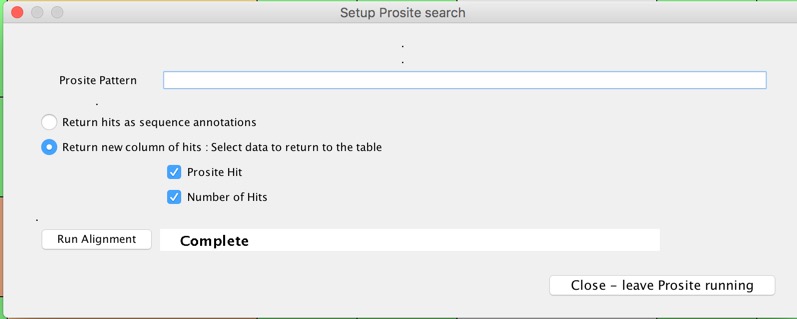
You can also do Prosite searches, in my brief trials these were effectively instantaneous.
This is certainly an interesting new area of data analysis, perhaps summed up by the current user guide help.
“This document is following a moving goal post, so may be a little behind the functionality. The biology material is also not core at this time.”
However I suspect this will soon be a core feature.
Dotmatics have released a video describing the new functionality
Last updated 19 November 2016
One thought on “Vortex does Biology”
Comments are closed.