t-distributed stochastic neighbor embedding (t-SNE) is a statistical method for visualizing high-dimensional data by giving each datapoint a location in a two or three-dimensional map. Whilst there are
Tag: rdkit
An interesting web app that fetches ChEMBL bioactivity data for a target (via UniProt ID), computes molecular descriptors, and trains a simple predictive model (regression, with
There is an interesting paper in Journal of Cheminformatics J Cheminform 17, 142 (2025). https://doi.org/10.1186/s13321-025-01094-1 Cheminformatics Microservice V3 aims to provide easily accessible and reproducible cheminformatics tools.
MolView – SMILES Preview is a Visual Studio Code extension that renders SMILES (Simplified Molecular Input Line Entry System) strings as molecular structure diagrams on
A while back I wrote a post on the problems with maintaining scientific software, this seemed to strike a chord and I got a lot
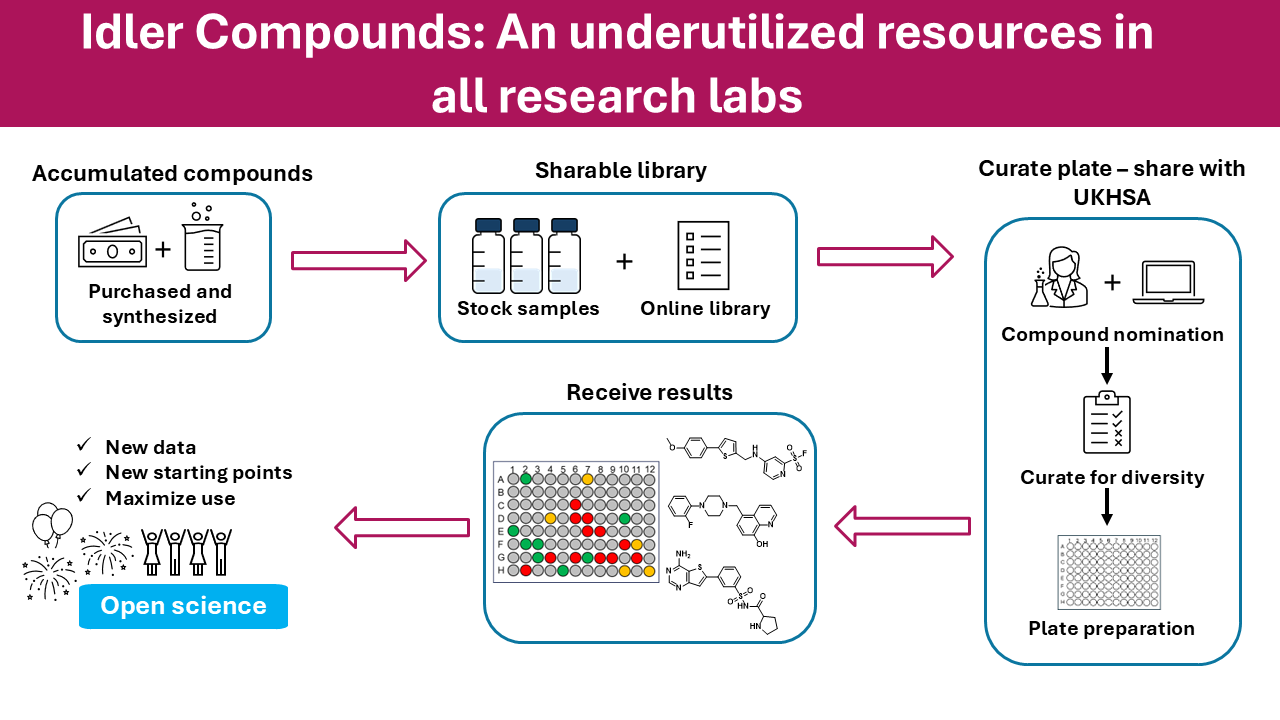
There was an interesting publication from the Todd group at UCL on Chemrxiv “Idler Compounds: A Simple Protocol for Openly Sharing Fridge Contents for Cross-Screening”

It is that time of the year when Royal Society of Chemistry membership forms arrive in the letterbox. Most people don’t join any of the
An interesting post from Nvidia. nvMolKit, is a standalone, GPU-accelerated library that delivers 1-4 orders of magnitude of acceleration across five key functions: Morgan Fingerprinting,
Whilst there are a number of chemistry aware spreadsheets Vortex, Stardrop, Datawarrior, InstantJChem, molsoft etc. many people still use Excel or Google sheets. Vexo is
PubChem is an invaluable source of information about 99 million molecules accessible via a website or programmatically. PubChem is an open chemistry database at the National Institutes
