t-distributed stochastic neighbor embedding (t-SNE) is a statistical method for visualizing high-dimensional data by giving each datapoint a location in a two or three-dimensional map. Whilst there are
Tag: python
An interesting web app that fetches ChEMBL bioactivity data for a target (via UniProt ID), computes molecular descriptors, and trains a simple predictive model (regression, with
Just a reminder the next Cambridge Cheminformatics meeting is on 18 February 2026, 4-5.30pm UK time, Hybrid (at the CCDC on Union Road, Cambridge/via Zoom)

Another addition to the superb MayaChemTools OpenFECalculateRelativeBindingFreeEnergySepTop.py to calculate Relative Binding Free Energy using a Separated Topologies (SepTop) approach. The script relies on the availability of OpenFE
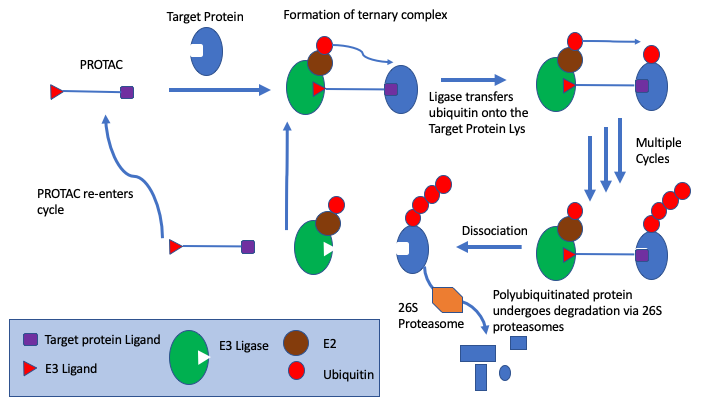
PROteolysis TArgeting Chimeras (PROTACs) technology provides an alternative to module biological function by specially using the ubiquitin proteasome system to induce degradation of the target
There is an interesting paper in Journal of Cheminformatics J Cheminform 17, 142 (2025). https://doi.org/10.1186/s13321-025-01094-1 Cheminformatics Microservice V3 aims to provide easily accessible and reproducible cheminformatics tools.
CNotebook provides chemistry visualization for Jupyter Notebooks and Marimo using the OpenEye Toolkits. Import the package and your molecular data will automatically render as chemical
MolView – SMILES Preview is a Visual Studio Code extension that renders SMILES (Simplified Molecular Input Line Entry System) strings as molecular structure diagrams on
A while back I wrote a post on the problems with maintaining scientific software, this seemed to strike a chord and I got a lot
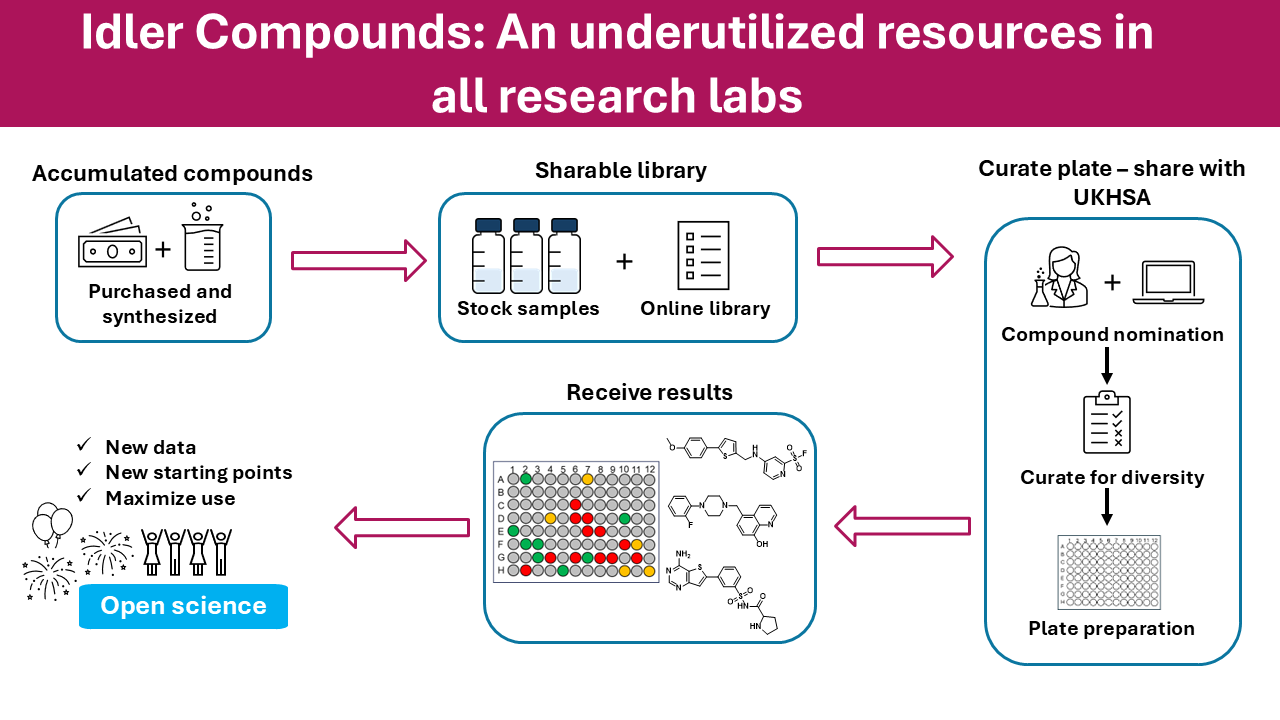
There was an interesting publication from the Todd group at UCL on Chemrxiv “Idler Compounds: A Simple Protocol for Openly Sharing Fridge Contents for Cross-Screening”
