In an earlier script I described the use of the ability to script multiple sub-structure searches using SMARTS. There are many occasions when this sort
Category: Vortex Scripts

ChEMBL is a manually curated chemical database of bioactive molecules . It is maintained by the European Bioinformatics Institute (EBI), of the European Molecular Biology Laboratory
One of the really neat features of the latest version of Vortex (> build 29622) is the ability to script multiple sub-structure searches using SMARTS.
Well things can change quickly at times, in the last tutorial I wrote.. Vortex has a limited capacity to render HTML, it is however a
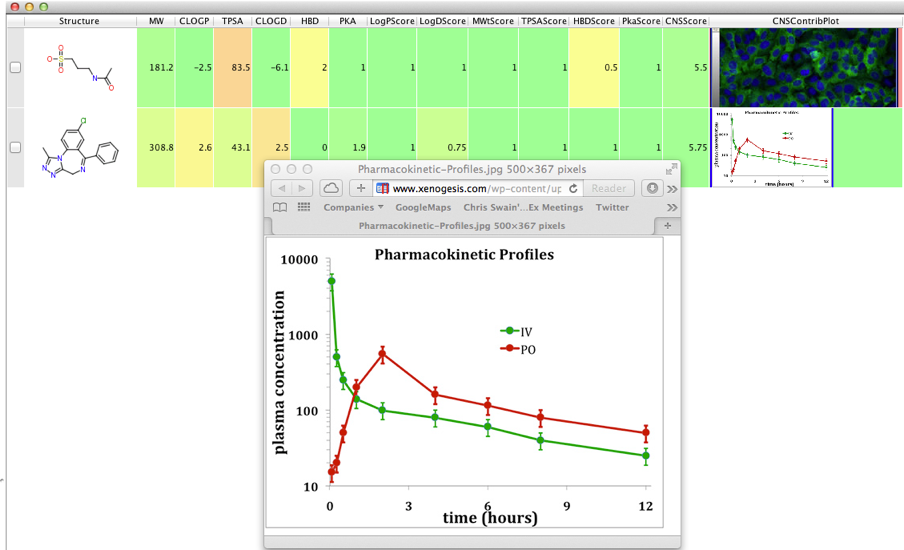
Vortex has a limited capacity to render HTML, it is however a very limited ability so there is no support for javascript or CSS but
It is clear from many publications that a number of physicochemical properties influence central nervous system (CNS) penetration and it is often possible to play
Un1Chem is a new web resource provided by the EBI, it is a ‘Unified Chemical Identifier’ system, designed to assist in the rapid cross-referencing of chemical

One of the new features in the latest version of MOE from Chemical Computing Group is the Listener. The MOE socket listener provides an alternative to MOE/web
About OCHEM OCHEM is a free open access site of annotated models and chemical data. OCHEM contains 1831772 experimental records for about 477 properties collected from

One of the new features in the latest version of MOE from Chemical Computing Group is the Listener. The MOE socket listener provides an alternative to MOE/web