CNotebook provides chemistry visualization for Jupyter Notebooks and Marimo using the OpenEye Toolkits. Import the package and your molecular data will automatically render as chemical
Author: chris
The next Cambridge Cheminformatics meeting is on 18 February 2026, 4-5.30pm UK time, Hybrid (at the CCDC on Union Road, Cambridge/via Zoom) More details of
MolView – SMILES Preview is a Visual Studio Code extension that renders SMILES (Simplified Molecular Input Line Entry System) strings as molecular structure diagrams on
A while back I wrote a post on the problems with maintaining scientific software, this seemed to strike a chord and I got a lot
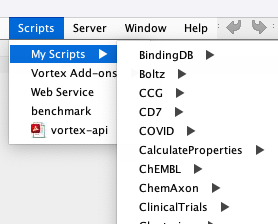
At least on a Mac the default place to store Vortex scripts is in the vortex folder ~/vortex/scripts Any folder/scripts you put in the vortex/scripts
Interesting post from ChEMBL team. We are excited to announce the start of the LIGAND-AI project, a 5-year project involving 18 partners to find ligands
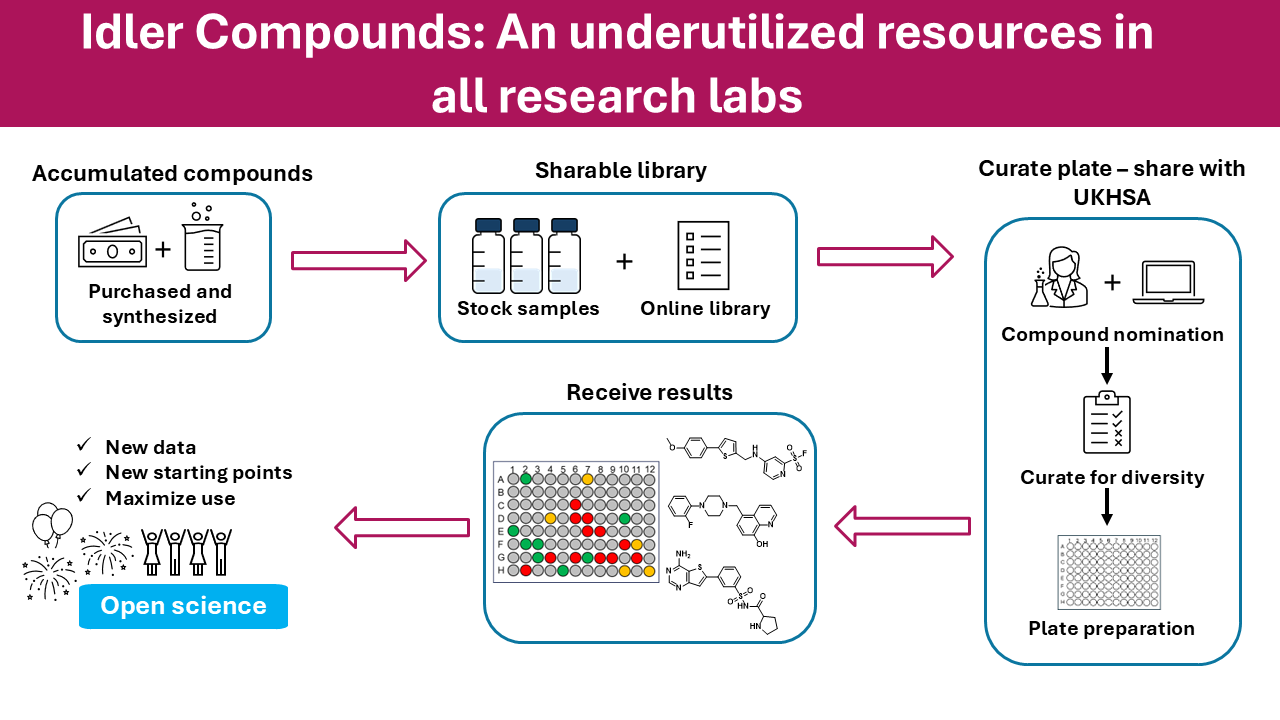
There was an interesting publication from the Todd group at UCL on Chemrxiv “Idler Compounds: A Simple Protocol for Openly Sharing Fridge Contents for Cross-Screening”

The life sciences sector is undergoing a profound transformation. Advances in automation, next-generation analytical technologies, and computational chemistry are redefining how research is conducted, making
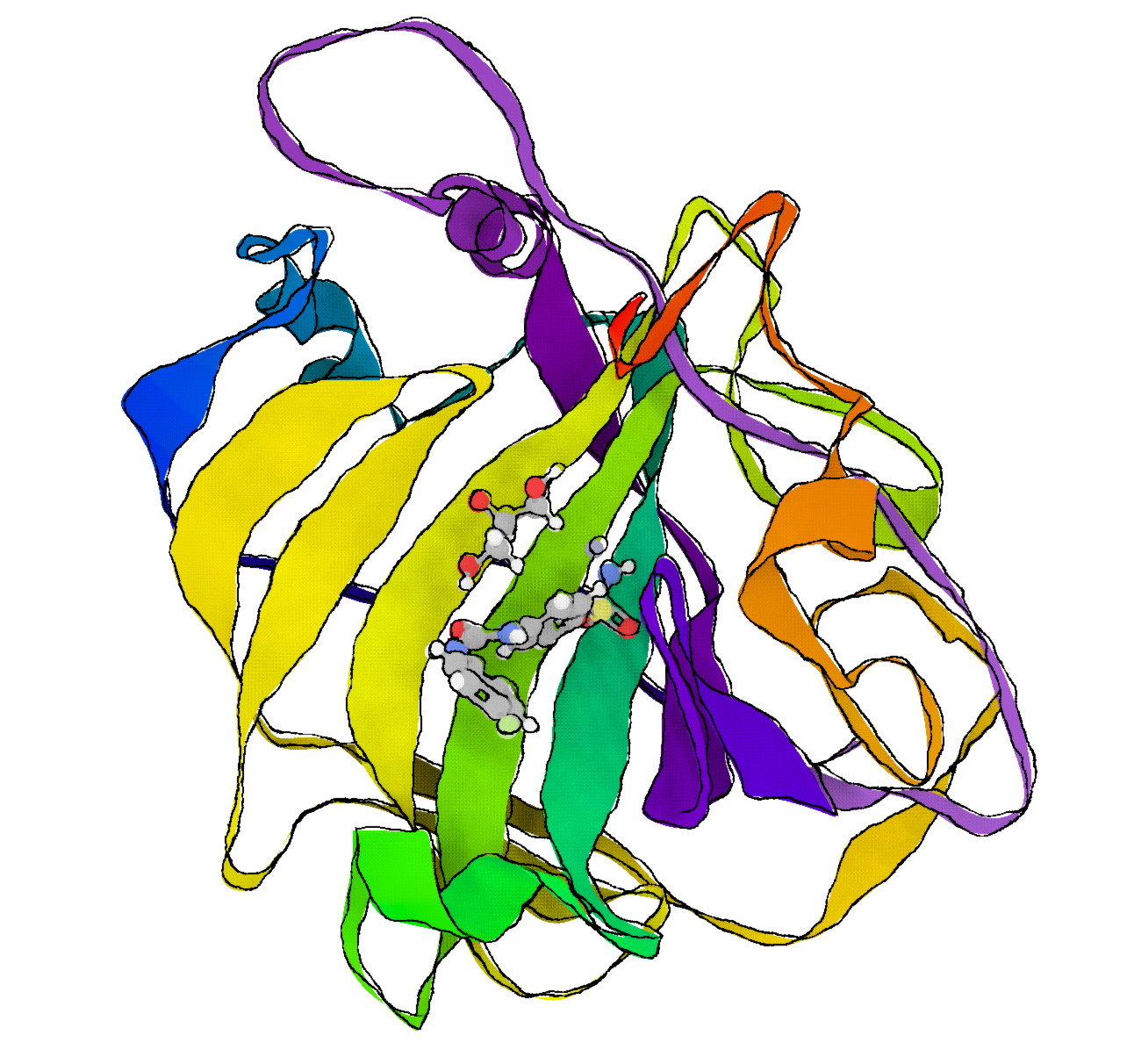
LigandExtractor can be used to find all ligands in a file from the PDB. In addition to finding all possible ligands, it annotates any problems
Next up Date: 5 February 2026 Lecture Theatre, Jeffrey Cheah Biomedical Centre, Puddicombe Way, Cambridge CB2 0AW. No need to register in advance – just