Sometimes when I’m comparing multiple datasets I end up with 10-20 different Vortex workspaces, for example if I’m comparing commercial screening collections from different vendors. I will calculated a variety of physicochemical properties, done clutter analysis, checked if each molecule has known biological activity in ChEMBL etc.

To share the results I often need to export to a sdf file, whilst it is straight forward to do this by hand it is a little tedious. Fortunately, I can write a simple script to do this automatically.
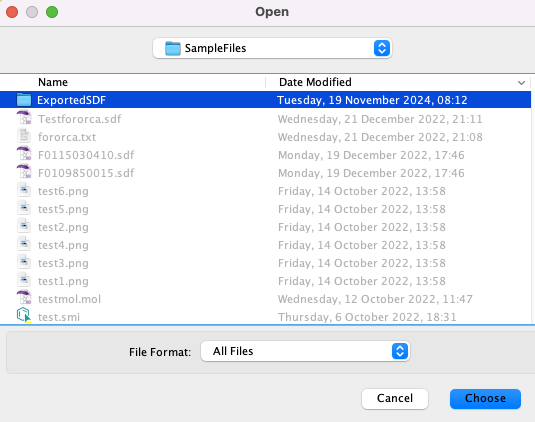
First open a dialog to choose the folder to save the exported sdf files in, then coerce the file to text.
Then get the number of workspaces, for each workspace get the name of the workspace (this will be used as the name of the exported sdf), the name of the table (only one table per workspace at present). Then export the table to the named sdf file. The script is shown below.
The Vortex Script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 |
#Created by Chris Swain www.macinchem.org # A script to export multiple workspaces to individual sdf files # Single table per workspace # Open a dialog to choose a folder to save sdf # vortex will keep track of the last folder you looked in etc folder = vortex.chooseDirectory() # Need to coerce file path txtFolder=folder.getAbsolutePath() #vortex.alert(txtFolder) num_workspaces = vortex.getWorkspaceCount() for n in range(0, num_workspaces): ws = vortex.getWorkspace(n) wsname = ws.name myvtable = ws.getTable() #ExportFile = "/Users/username/Desktop/Temp/" + wsname ExportFile = txtFolder +"/" + wsname myvtable.exportAsSdf(ExportFile) |
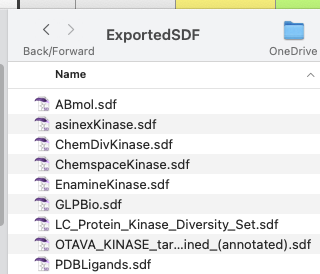
The result is a folder containing the exported sdf files.