The two previous vortex scripts used the PBD id to download information about a biomolecular structure structure from the Worldwide Protein Data Bank.
The second script creates a column in the Vortex workspace containing the identifiers for ligands pulled from the ligand information (This api call provides a a list of modelled instances of ligands, i.e. ‘bound’ molecules that are not waters.).
Vortex can display chemical structures as well as alphanumeric information so this additional script retrieves the corresponding ligand structures as SMILES strings which are then rendered by Vortex
ChemID2SMILES vortex script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 |
#Use PDB Ligand ID to find Ligand SMILES #Authored by Chris Swain (http://www.macinchem.org) #Uses PDBe #https://www.ebi.ac.uk/pdbe/api/pdb/compound/summary/ATP # Python imports import urllib2 import urllib from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os import re input_label = swing.JLabel("Ligands column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) # Get column containing Chem id ret = vortex.showInDialog(panel, "Choose Ligand column") if ret == vortex.OK: input_idx = input_cb.getSelectedIndex() if input_idx == 0: vortex.alert("you must choose a column") else: col = vtable.getColumn(input_idx - 1) #Format of query url #https://www.ebi.ac.uk/pdbe/api/pdb/compound/summary/ATP rows = vtable.getRealRowCount() for r in range(0, int(rows)): chemidstring = col.getValueAsString(r) chemidlist =re.split(r',',chemidstring) chemidlist = list(set(chemidlist)) #get unique ID #vortex.alert(chemidlist) for x in range(len(chemidlist)): chemid = chemidlist[x] api_url = "https://www.ebi.ac.uk/pdbe/api/pdb/compound/summary/" + chemid #vortex.alert(api_url) try: molecule_record = urllib2.urlopen(api_url).read() except urllib2.HTTPError: continue j = json.loads(molecule_record) TheSMILES = str(j[chemid][0]["smiles"][0]["name"]) colname = "SMILES" + str(x) colsmiles= vtable.findColumnWithName(colname, 1) colsmiles.setValueFromString(r, TheSMILES) vtable.fireTableStructureChanged() #Data format #{"ATP":[{"formula":"C10 H16 N5 O13 P3","creation_date":"19990708","revision_date":"20110604","name":"ADENOSINE-5'-TRIPHOSPHATE","weight":507.181,"formal_charge":0,"one_letter_code":"X","chembl_id":"CHEMBL14249","chebi_id":15422,"subcomponent_occurrences":{},"inchi":"InChI=1S/C10H16N5O13P3/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(26-10)1-25-30(21,22)28-31(23,24)27-29(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H,23,24)(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1","inchi_key":"ZKHQWZAMYRWXGA-KQYNXXCUSA-N","systematic_names":[{"program":"ACDLabs","version":"10.04","name":"adenosine 5'-(tetrahydrogen triphosphate)"},{"program":"OpenEye OEToolkits","version":"1.5.0","name":"[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate"}],"smiles":[{"program":"CACTVS","version":"3.341","name":"Nc1ncnc2n(cnc12)[C@@H]3O[C@H](CO[P@](O)(=O)O[P@@](O)(=O)O[P](O)(O)=O)[C@@H](O)[C@H]3O"},{"program":"OpenEye OEToolkits","version":"1.5.0","name":"c1nc(c2c(n1)n(cn2)[C@H]3[C@@H]([C@@H]([C@H](O3)CO[P@@](=O)(O)O[P@](=O)(O)OP(=O)(O)O)O)O)N"}],"stereoisomers":[{"chem_comp_id":"HEJ","name":"9-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-amine"}]}]} |
The first part of the script is the usual boilerplate, the dialog box asks the user to identify the column containing the ligand ID. Since there may be multiple ligands, and multiple instances of a specific ligand we convert to a set and then back to list to get a list of unique ID.
|
1 |
chemidlist = list(set(chemidlist)) |
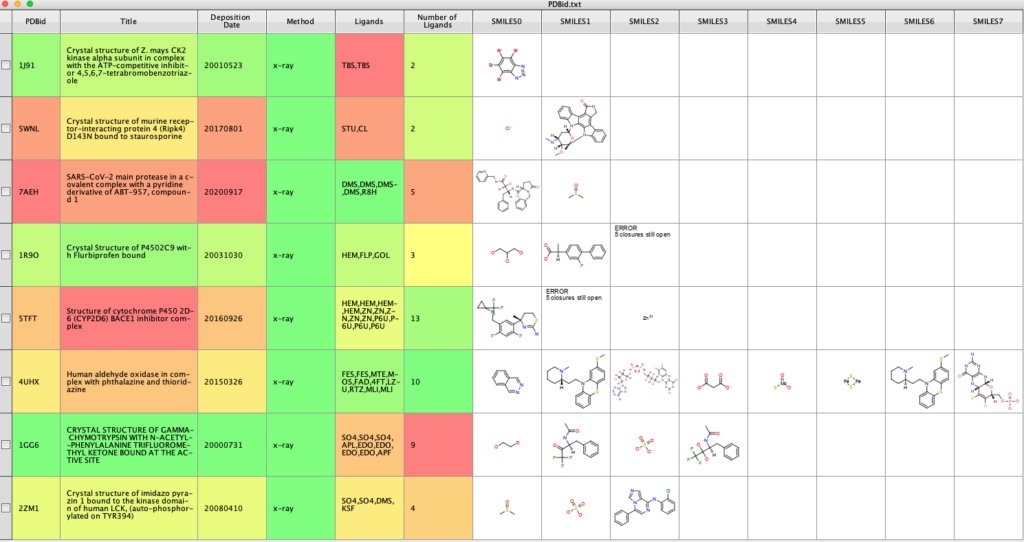
This list is then used to create the api_url to use the web service to access the molecular information about each ligand. The returned json is then parsed to extract all the SMILES strings and the individual SMILES copied into separate columns. The result is shown below. The json also includes number of molecular properties that could also be exported.
The script can be downloaded here.https://macinchem.org/wp-content/uploads/2025/04/ChemID2SMILES.vpy_.zip
