In a previous post I illustrated how to download PubChem and create a local searchable database using a Jupyter notebook. I also included a vortex/python script to search for PubChem ID from a InChiKey. A couple of readers have asked if I could give more examples, but using a smaller database since the PubChem sqlite database with fingerprints is over 450 GB!
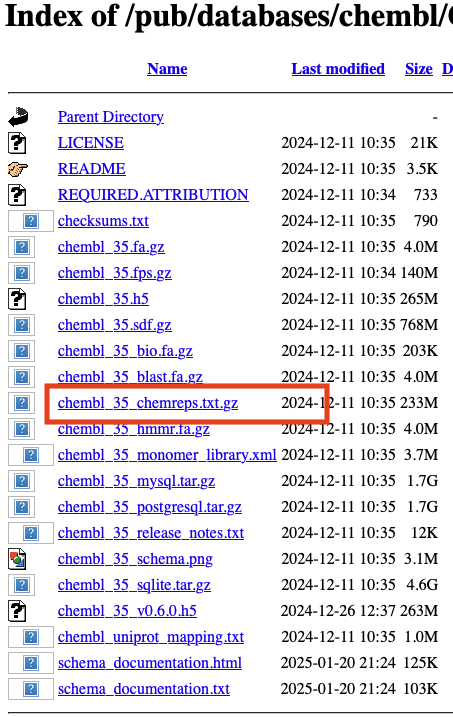
So instead I’ve created a Jupyter notebook that uses the latest version of ChEMBL database that can be downloaded from their ftp site, the file you need is highlighted below. https://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_35/
The Jupyter notebook is shown below and can be downloaded here
The Jupyter notebook can be used to generate the chembl.sqlite database and also includes various cells that show options for querying the database. These can be simply modified to python scripts that can be called from the command line or accessed via a Vortex script.
Search using InChiKey
The python script below takes as input an inChiKey and returns the chembl_id.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
#!/usr/bin/env python # coding: utf-8 import sqlite3 import io import os import sys targetInChiKey = sys.argv[1] connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBL/chembldb.sqlite") #using external SSD #connection = sqlite3.connect("/Volumes/ExternaLSSD/Projects/ChEMBL/chembldb.sqlite") cursor = connection.cursor() #targetInChiKey = "WPQAOGZTDKTBHI-UHFFFAOYSA-N" targetID = cursor.execute("SELECT chembl_id FROM chembl WHERE INCHIKEY= ?", (targetInChiKey,),).fetchall() if not targetID: ReturnedData = "Not Found" else: ReturnedData = targetID[0][0] print(ReturnedData) # python /Users/chrisswain/Projects/ChEMBL/searchinchikey.py WPQAOGZTDKTBHI-UHFFFAOYSA-N |
We can test this using the command line.
|
1 2 |
% python /Users/chrisswain/Projects/ChEMBL/searchinchikey.py ZGLJYEZERNATQV-UHFFFAOYSA-N CHEMBL3318011 |
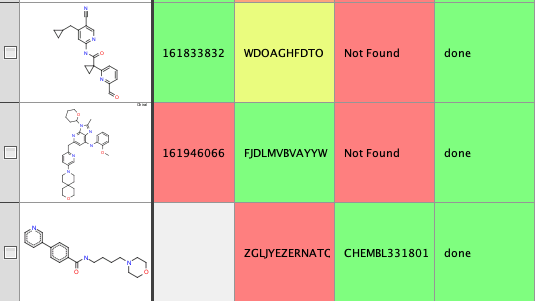
And we can access this using a Vortex script, the first part of the script asks the user to select the column containing the InChiKeys, subprocess is then used to generate the query and collect the response. The response is then entered into the Vortex workspace.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 |
#Use InChiKey to search Local copy of ChEMBL to get ID #Authored by Chris Swain (http://www.macinchem.org) #All rights reserved. # Python imports import urllib2 import urllib import subprocess from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os myhome = os.getenv('HOME') input_label = swing.JLabel("InChikey column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) ret = vortex.showInDialog(panel, "Choose InChiKey column") if ret == vortex.OK: input_idx = input_cb.getSelectedIndex() if input_idx == 0: vortex.alert("you must choose a column") else: chosen_col = vtable.getColumn(input_idx - 1) colChEMBL = vtable.findColumnWithName('ChEMBL ID', 1) coldone = vtable.findColumnWithName('Done', 1) rows = vtable.getRealRowCount() for r in range(0, int(rows)): inchikey_id = chosen_col.getValueAsString(r) coldone.setValueFromString(r, "done") errorText = "error" # python /Users/chrisswain/Projects/ChEMBL/searchinchikey.py WPQAOGZTDKTBHI-UHFFFAOYSA-N #vortex.alert('INChiKey = '+inchikey_id) p = subprocess.Popen(['/Users/chrisswain/miniconda3/bin/python', '/Users/chrisswain/Projects/ChEMBL/searchinchikey.py', inchikey_id], stdout=subprocess.PIPE) output = p.communicate()[0] #vortex.alert(output) colChEMBL.setValueFromString(r, output) vtable.fireTableStructureChanged() |
The result is shown below.
ChEMBL ID to SMILES
If you have a list of ChEMBL ID and you want to add the structures the script should help.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 |
#!/usr/bin/env python # coding: utf-8 import sqlite3 import io import os import sys from rdkit import Chem targetInChiKey = sys.argv[1] connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBL/chembldb.sqlite") #using external SSD #connection = sqlite3.connect("/Volumes/ExternaLSSD/Projects/ChEMBL/chembldb.sqlite") cursor = connection.cursor() targetID = cursor.execute("SELECT SMILES FROM chembl WHERE chembl_id= ?", (targetInChiKey,),).fetchall() if not targetID: ReturnedData = "" else: ReturnedData = targetID[0][0] print(ReturnedData) |
Again this can be tested from the command line, and accessed from a Vortex script in a similar manner to before.
|
1 2 |
% python /Users/chrisswain/Projects/ChEMBL/CHEMBLID2SMILES.py CHEMBL1318887 Clc1ccc(-c2ccccc2)cn1 |
Count of molecules containing a substructure
If you have a list of molecules and you want to know how many times they appear as a substructure in ChEMBL. In this case we are using the chemicalite extension so after connecting to the database chemicalite is loaded. The SQL query is created and run.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 |
#!/usr/bin/env python # coding: utf-8 import sqlite3 import io import os import sys from rdkit import Chem targetSMILES = sys.argv[1] connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBL/chembldb.sqlite") #using external SSD #connection = sqlite3.connect("/Volumes/ExternaLSSD/Projects/ChEMBL/chembldb.sqlite") cursor = connection.cursor() connection.enable_load_extension(True) connection.load_extension('chemicalite') connection.enable_load_extension(False) #targetID = cursor.execute("SELECT SMILES FROM chembl WHERE chembl_id= ?", (targetInChiKey,),).fetchall() count = connection.execute("SELECT COUNT(*) FROM chembl, str_idx_chembl_molecule AS idx " + "WHERE chembl.id = idx.id AND mol_is_substruct(chembl.molecule, mol_from_smiles('" + targetSMILES + " ')) " + "AND idx.id MATCH rdtree_subset(mol_pattern_bfp(mol_from_smiles('" + targetSMILES + "'), 2048))").fetchall()[0][0] if not count: ReturnedData = "" else: ReturnedData = count print(ReturnedData) |
We can test this from the command line but there are a couple of things to watch. the python environment used to create the database needs to be used. In addition, SMILES can contain characters that might be interpreted incorrectly so they should be enclosed with quotes.
|
1 2 |
% /Users/chrisswain/miniconda3/envs/rdkitenv/bin/python3.11 /Users/chrisswain/Projects/ChEMBL/CountSSS.py "Clc1ccc(-c2ccccc2)cn1" 226 |
The Vortex script is shown below. The first part gets the SMILES from the structure and then we generate the query, making sure to put the SMILES in quotes.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
#Use SMILES to substructure search Local copy of ChEMBL to get count #Authored by Chris Swain (http://www.macinchem.org) #All rights reserved. from javax.swing import JComboBox, JPanel, JLabel, JSpinner, SpinnerNumberModel, JCheckBox import com.dotmatics.vortex.util.Layout as layout import itertools # Python imports import urllib2 import urllib import subprocess from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os myhome = os.getenv('HOME') #for testing #theSMILES = "C1=CC=C(C=C1)C1=CC=CN=C1" colNumSim = vtable.findColumnWithName('ChEMBL SSS', 1) coldone = vtable.findColumnWithName('Done', 1) rows = vtable.getRealRowCount() for r in range(0, int(rows)): coldone.setValueFromString(r, "done") theSMILES = vortex.getMolProperty(vtable.getMolFileManager().getMolFileAtRow(r), 'SMILES') #/Users/chrisswain/miniconda3/envs/rdkitenv/bin/python3.11 /Users/chrisswain/Projects/ChEMBL/Countsimilar.py "C1=CC=C(C=C1)C1=CC=CN=C1" p = subprocess.Popen(['/Users/chrisswain/miniconda3/envs/rdkitenv/bin/python3.11', '/Users/chrisswain/Projects/ChEMBL/CountSSS.py', theSMILES], stdout=subprocess.PIPE) output = p.communicate()[0] colNumSim.setValueFromString(r, output) vtable.fireTableStructureChanged() |
Count of similar molecules
An alternative search might to try and find how many similar molecules there are. The python script is shown below, there are a couple of points to note. Again the chemiclite extension needs to be loaded. Command-line arguments are STRINGS so the threshold value (Tanimoto similarity) needs to be converted to a FLOAT.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 |
#!/usr/bin/env python # coding: utf-8 import sqlite3 import io import os import sys from rdkit import Chem targetSMILES = sys.argv[1] #targetSMILES = "O=c1ccc(CCCc2ccccc2)cn1O" threshold = float(sys.argv[2]) #convert argv to float connection = sqlite3.connect("/Users/chrisswain/Projects/ChEMBL/chembldb.sqlite") #using external SSD #connection = sqlite3.connect("/Volumes/ExternaLSSD/Projects/ChEMBL/chembldb.sqlite") cursor = connection.cursor() connection.enable_load_extension(True) connection.load_extension('chemicalite') connection.enable_load_extension(False) rs = connection.execute( "SELECT c.chembl_id, mol_to_smiles(c.molecule), " + "bfp_tanimoto(mol_morgan_bfp(c.molecule, 2, 1024), " " mol_morgan_bfp(mol_from_smiles(?1), 2, 1024)) as t " + "FROM " + "chembl as c JOIN morgan_idx_chembl_molecule as idx USING(id) " + "WHERE " + "idx.id MATCH rdtree_tanimoto(mol_morgan_bfp(mol_from_smiles(?1), 2, 1024), ?2) " + "ORDER BY t DESC", (targetSMILES, threshold)).fetchall() count = len(rs) if not count: ReturnedData = "" else: ReturnedData = count print(ReturnedData) |
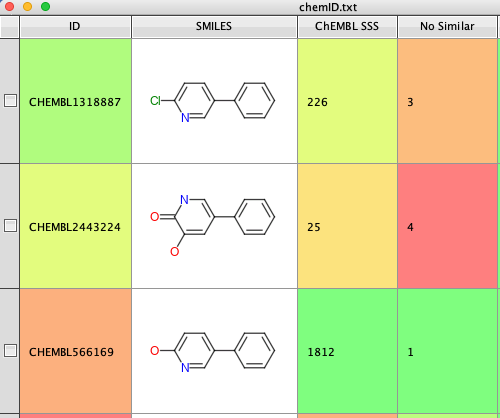
The results of these three scripts are shown below, starting with a list of ChEMBL ID, first the SMILES string is added, then the results of the substructure search and then the number of similar (Tanimoto 0.6) are added
The python and Vortex scripts can be downloaded here
I put the python scripts in the same folder as the chembl.sqlite database, and the Vortex scripts into a “ChEMBL” folder I created inside the Vortex scripts folder. You will need to edit the paths within the scripts for your machine.
Some of the more eagle-eyed will have noticed the commented out line in the python code shown below, given that the sqlite databases can be quite large I also tested having them on an external drive. I tried out various options and ended up buying an ACASIS 40Gbps M.2 NVMe SSD Enclosure with Crucial P3 Plus 4TB SSD (Thunderbolt).
|
1 |
connection = sqlite3.connect("/Volumes/ExternaLSSD/Projects/ChEMBL/chembldb.sqlite") |
Using the external drive was slightly slower but I’ve not done extensive testing