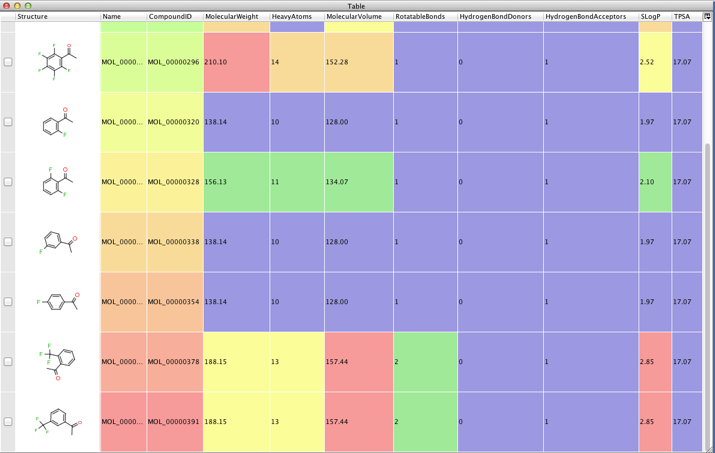
In previous scripts I’ve shown how to use a couple of command line applications to calculate physicochemical properties. In this tutorial we will use a PERL script that is part of the excellent MayaChemTools.
MayaChemTools is a growing collection of Perl scripts, modules, and classes to support day-to-day computational discovery needs The current release of MayaChemTools provides command line scripts for the following tasks:
* Manipulation of SD, CSV/TSV, Sequence/Alignments, and PDB files
* Analysis of data in SD, CSV/TSV, and Sequence/Alignments files
* Information about data in SD, CSV/TSV, Sequence/Alignments, PDB, and fingerprints files
* Exporting data from Oracle and MySQL tables into text files
* Properties of periodic table elements, amino acids, and nucleic acids
* Elemental analysis
* Generation of fingerprints corresponding to atom neighbourhoods, atom types, E-state indicies, extended connectivity, MACCS keys, path lengths, topological atom pairs, topological atom triplets, topological atom torsions, topological pharmacophore atom pairs, and topological pharmacophore atom triplets
* Generation of fingerprints with atom types corresponding to atomic invariants, DREIDING, E-state, functional class, MMFF94, SLogP, SYBYL, TPSA and UFF
* Calculation of similarity matrices using a variety of similarity and distance coefficients
* Calculation of physicochemical properties including rotatable bonds, van der Waals molecular volume, hydrogen bond donors and acceptors, logP (SLogP), molar refractivity (SMR), topological polar surface area (TPSA), molecular complexity, and so on
* Similarity searching using fingerprints
MayaChemTools is free software; you can redistribute it and/or modify it under the terms of the GNU LGPL as published by the Free Software Foundation.
After downloading you need to decide where you are going to put the mayachemtools folder, I put is in /usr/local, you then need to add /bin to your PATH environment variable. Check to make sure PATH doesn’t contain multiple entries for MayaChemtools package, and all *.pl files in /bin are executable using the following command.
chmod +x filename
There over 50 perl scripts in this valuable collection but we will be using just one, CalculatePhysicochemicalProperties.pl. Currently the following properties are supported MolecularWeight, ExactMass, HeavyAtoms, Rings, AromaticRings, van der Waals MolecularVolume, RotatableBonds, HydrogenBondDonors, HydrogenBondAcceptors, LogP and Molar Refractivity (SLogP and SMR), Topological Polar, Surface Area (TPSA), Fraction of SP3 carbons (Fsp3Carbons) and SP3 carbons (Sp3Carbons), MolecularComplexity.
The documentation gives full details of the options but we will be using the command shown below, where you will need to replace “username” with your username to test it.
/usr/local/mayachemtools/bin/CalculatePhysicochemicalProperties.pl --CompoundIDMode MolName --CompoundIDLabel CompoundID --OutDelim tab -o -q No -r /Users/username/Desktop/MayaOutput /Users/username/Desktop/Output.sdf
This PERL script calculates a number of properties for the molecules in the input file and writes the results to /Users/username/Desktop/MayaOutput.tsv as tab separated values without quotes, the first line of the file containing the property names.
In the Vortex script we don’t want to hard code the full path to the output file we use the code below to get the path to the desktop and then append the filename, if you want to save the file elsewhere you will need to modify this part of the script.
import os
# get path to desktop
mydesk=os.path.join(os.path.expanduser("~"), "Desktop")
outputfile=(mydesk + '/MayaOutput')
Note the perl script automatically appends the file suffix to the output file so we don’t add it here. The next part of the script runs the command creating the output, it took me a little time to to get this part of the script functioning correctly, it seems each white space delimited argument need to be a separate string in the subprocess call. Running th script stepwise in the Vortex console app certainly helped trouble-shooting.
Then when we read the resulting file, as you can see we have to append the the suffix in this case.
# Read output file
f = open(mydesk + '/MayaOutput.tsv', 'r')
output=f.read()
Having got the data we then add it to the table we parse the data as described in previous scripts, and add it to the data table.

The Vortex script
import sys
#Uncomment the following 2 lines if running in console
#vortex = console.vortex
#vtable = console.vtable
sys.path.append(vortex.getVortexFolder() + '/modules/jythonlib')
import subprocess
# Get the path to the currently open sdf file
sdfFile = vortex.getFileForPropertyCalculation(vtable)
import os
# get path to desktop
mydesk=os.path.join(os.path.expanduser("~"), "Desktop")
outputfile=(mydesk + '/MayaOutput')
# Run mayachemtools on the file
# /usr/local/mayachemtools/bin/CalculatePhysicochemicalProperties.pl --CompoundIDMode MolName --CompoundIDLabel CompoundID --OutDelim tab -o -r SampleOutput /Users/usename/Desktop/ChemicalStructures/acetophenones.sdf;cat SampleOutput.tsv
p =subprocess.Popen(['/usr/local/mayachemtools/bin/CalculatePhysicochemicalProperties.pl', '--CompoundIDMode', 'MolName' , '--CompoundIDLabel','CompoundID', '--OutDelim', 'tab', '-o', '-q', 'No', '-r', outputfile, sdfFile],stdout=subprocess.PIPE)
output = p.communicate()[0]
# Read output file
f = open(mydesk + '/MayaOutput.tsv', 'r')
output=f.read()
# Create new columns in table if needed
lines = output.split('\n')
colName = lines[0].split('\t')
for c in colName:
column = vtable.findColumnWithName(c, 1)
vtable.fireTableStructureChanged()
keys = []
for i in lines:
words = i.split('\t')
if len(words) == 2:
keys.append(words[0])
# Parse the output
rows = lines[1:len(lines)]
for r in range(0, vtable.getRealRowCount()):
vals = rows[r].split('\t')
for j in range(0, len(vals)):
column = vtable.findColumnWithName(colName[j], 0)
column.setValueFromString(r, vals[j])
The script can be downloaded from here