The Open Source Malaria project is trying a different approach to curing malaria. Guided by open source principles, everything is open and anyone can contribute. To date a lot of people around the world have made contributions and the project is at a very exciting stage. Whilst everyone can see the compounds that have been made and the biological data, it is often spread over multiple web pages and can be tricky to link molecule with identifier with data. Over the last couple of months a significant effort has been put into populating a spreadsheet with all the information.
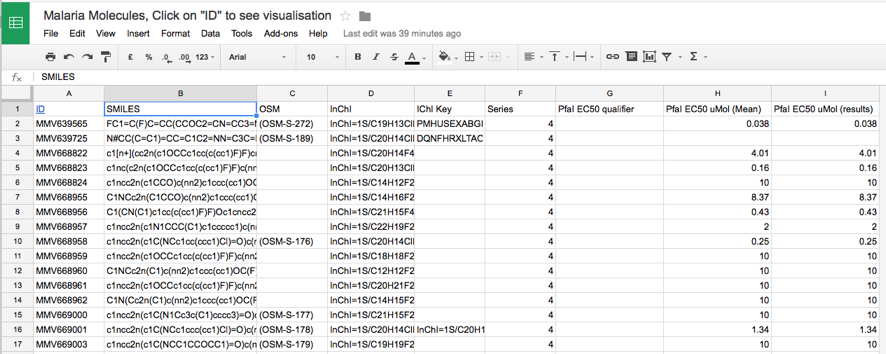
The plan is that all new molecules will be added to the spreadsheet and new assays will added as additional columns. Storing the structures in a text format like SMILES provides a compact and efficient way to store molecular information which does not require any specials software. Whilst this provides a useful repository it is not particularly helpful for the chemists who would actually prefer to see the structures of the molecules.
In collaboration with Luc Patiny at http://www.cheminfo.org/ we have been able to provide a visualiser that pulls data directly from the spreadsheet. This currently requires Google Chrome. Link to visualiser. This also calculates a number of physicochemical properties on the fly.
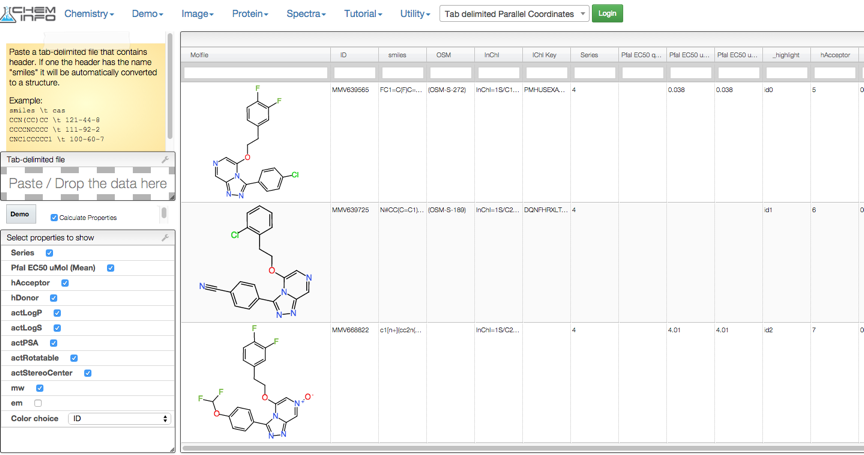
Whilst this is very, very useful for viewing results it is not ideal for trying to build predictive models. Vortexis a chemically intelligent data analysis and visualisation platform. This script provides a one-click access to the OSM data and creates a new workspace containing the data, and since it is linked to the live spreadsheet you will always have access to the latest data.
OSMdata Vortex script
The first part of the script imports the data from the google spreadsheet as tab separated values, we then store the data as an array in list1.
We can then get the column names by parsing the first line of list1.
We then get the data by parsing each line of list1 starting at the second line.
Finally we create a new workspace.
# Python imports
import urllib2
import urllib
import csv
import sys
from com.xhaus.jyson import JysonCodec as json
# Vortex imports
import com.dotmatics.vortex.util.Util as Util
import com.dotmatics.vortex.mol2img.jni.genImage as genImage
import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img
import com.dotmatics.vortex.table.VortexTableModel as vtm
import jarray
import binascii
import string
import os
# Example search string
# http://docs.google.com/spreadsheets/d/1Rvy6OiM291d1GN_cyT6eSw_C3lSuJ1jaR7AJa8hgGsc/export?format=tsv
mystr = "http://docs.google.com/spreadsheets/d/1Rvy6OiM291d1GN_cyT6eSw_C3lSuJ1jaR7AJa8hgGsc/export?format=tsv"
myreturn = urllib2.urlopen(mystr).read()
list1 = myreturn.split('\n')
TableName = "OSMData"
# Get column names
column_names = list1[0].split('\t')
rows = []
for i in list1[1:]:
row = i.split('\t')
rows.append(row)
arrayToWorkspace(rows, column_names, TableName)
The results are shown below.
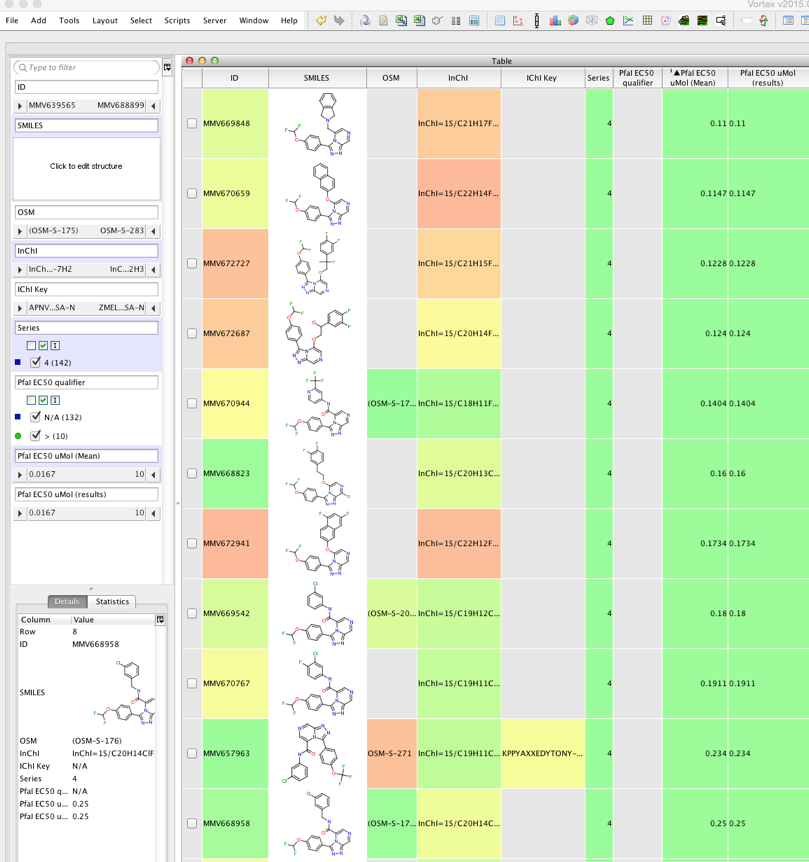
The script can be downloaded from here why not give it a try and then contribute your findings and suggestions to the Open Source Malaria project.
Page Updated 24 June 2015