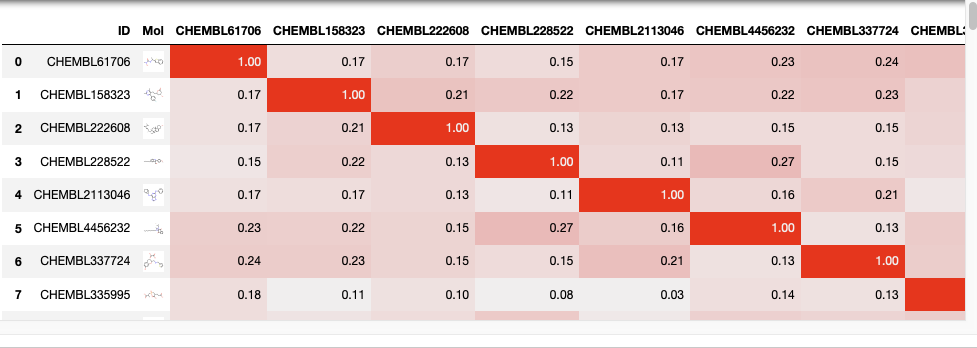
I’m sometimes asked for a tool to compare the similarity of a list of molecules with every other molecule in the list. I suspect there
Tag: rdkit

Registration for the 2024 RDKit UGM, being held from 11-13 September at the ETH in Zurich, Switzerland, is now open. https://www.eventbrite.com/e/13th-rdkit-ugm-2024-tickets-860637719587 The UGM will be
This is very cool. Clipboard-to-SMILES Converter (C2SC), is a macOS application that directly converts molecular structures from the clipboard. The app focuses on seamlessly converting
Another tutorial from Greg Landrum, focussing on how to take advantage of modern multithreaded CPUs. https://greglandrum.github.io/rdkit-blog/posts/2024-02-11-more-multithreading.html Looking at Generating fingerprints, Molecular standardization, Conformer generation, RMSD
mmpdb 3.0, released May 2023, merged three development tracks: – create and query 1-cut med chem transformations as described in Awale et al., The Playbooks
Scikit-Mol originated during an RDKit hackathon and has gradually evolved into a tool that I looks extremely useful. Scikit-Mol is related to Scikit-learn and molecules,
Another script from the fantastic MayaChemTools collection created by Manish Sud. A new command line script named VinaPerformDocking.py. The script relies on the presence of AutoDock Vina, Meeko,
If you need to display molecules and associated data in a grid then Jeremy Monat’s MolsMatrixToGridImage is exactly what you need. To underline just how
As virtual libraries get ever larger the challenges of virtual screening get larger. Whilst docking into a target protein is a very popular and successful

2023_09_1 (Q3 2023) Release https://github.com/rdkit/rdkit/releases/tag/Release_2023_09_1 Highlights Plus lots of other changes and enhancements