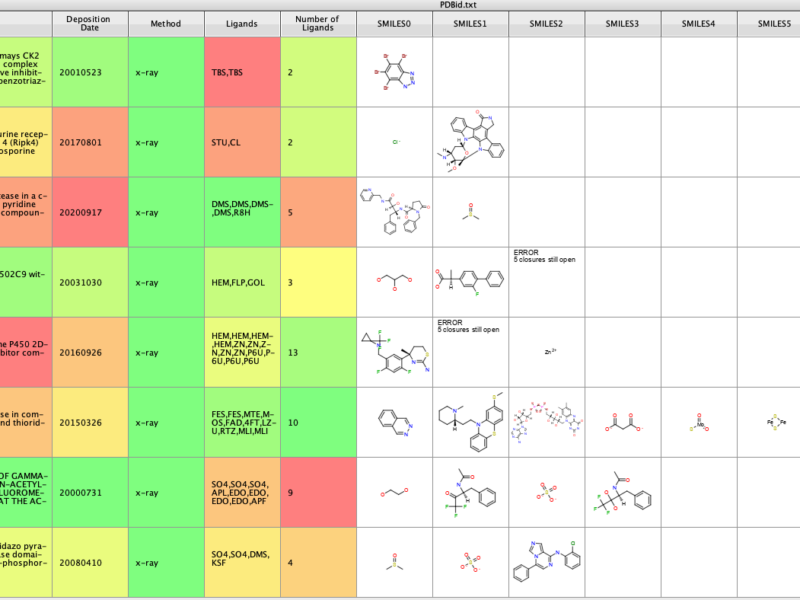
The previous scripts have allowed the user to get more information about a PDB entry and to import the ligand structures. This script allows the user to identify any PDB structures associated with a Uniprot ID (https://www.uniprot.org).
Uniprot2PDB
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 |
#Use Uniprot accession id to find PDB structures #Authored by Chris Swain (http://www.macinchem.org) #All rights reserved. #https://www.ebi.ac.uk/pdbe/api/mappings/P50225 # Python imports import urllib2 import urllib from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os input_label = swing.JLabel("Uniprot column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) ret = vortex.showInDialog(panel, "Choose Uniprot column") if ret == vortex.OK: input_idx = input_cb.getSelectedIndex() if input_idx == 0: vortex.alert("you must choose a column") else: col = vtable.getColumn(input_idx - 1) colpdbid = vtable.findColumnWithName('PDB', 1) colpdbNo = vtable.findColumnWithName('Num PDB entries', 1) rows = vtable.getRealRowCount() for r in range(0, int(rows)): uniprotid = col.getValueAsString(r) try: api_url = "https://www.ebi.ac.uk/pdbe/api/mappings/" + uniprotid #api_url = "https://www.ebi.ac.uk/pdbe/api/mappings/P50225" molecule_record = urllib2.urlopen(api_url).read() #vortex.alert(molecule_record) j = json.loads(molecule_record) ThePDBs = j[uniprotid]["PDB"].keys() #vortex.alert(str(ThePDBs)) pdbString = ','.join(ThePDBs) #vortex.alert(pdbString) value = len(ThePDBs) #vortex.alert(value) colpdbid.setValueFromString(r, pdbString) colpdbNo.setValueFromString(r, str(value)) except: error = "Not Found" vtable.fireTableStructureChanged() #Data format "P50225":{"PDB":{"3u3k":[{"entity_id":1,"chain_id":"B","struct_asym_id":"B","unp_start":1,"unp_end":295,"start":{"residue_number":21,"author_residue_number":null,"author_insertion_code":""},"end":{"residue_number":315,"author_residue_number":295,"author_insertion_code":""}},{"entity_id":1,"chain_id":"A","struct_asym_id":"A","unp_start":1,"unp_end":295,"start":{"residue_number":21,"author_residue_number":null,"author_insertion_code":""},"end":{"residue_number":315,"author_residue_number":295,"author_insertion_code":""}}],"1z28":[{"entity_id":1,"chain_id":"A","struct_asym_id":"A","unp_start":1,"unp_end":295,"start":{"residue_number":1,"author_residue_number":null,"author_insertion_code":""} |
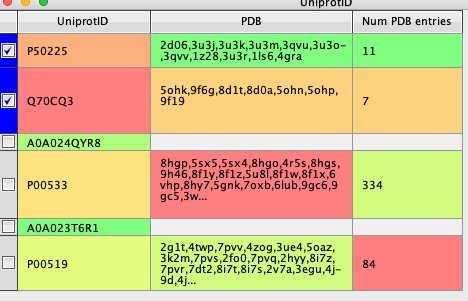
The results is shown in the image below, there is a list of PDB id and a count of how many entries.
The script can be downloaded here