TabPFN is a foundation model trained on around 130,000,000 synthetically generated datasets that mimic “real world” tabular data. These datasets sampled dataset size and number of features, both classification and regression tasks, and Gaussian noise was added to mimic real-world complexities. This can then be used to build models for small- to medium-sized datasets with up to 10,000 samples and 500 features and is claimed to be superior to other methods.
Here we present the Tabular Prior-data Fitted Network (TabPFN), a tabular foundation model that outperforms all previous methods on datasets with up to 10,000 samples by a wide margin, using substantially less training time.
I previously posted details of testing TabPFN on Apple Silicon and found it performed interestingly well. Unfortunately, the datasets used were not ideal and required significant tidying up before they could be used. The variable quality of datasets has been highlighted by Pat Walters on the Practical Cheminformatics blog.
We shouldn’t consider something a standard for the field simply because everyone blindly uses it. It should also be noted that while the points I make below primarily focus on MoleculeNet, other widely used benchmarks like the Therapeutic Data Commons (TDC) are equally flawed.
To address this Polaris have established a novel, industry‑certified standard to foster the development of impactful methods in AI-based drug discovery. There are a collection of datasets that can be used to build ML/AI models and in this post the biogen/adme-fang-v1 was used. This contains several datasets including liver microsome clearance, solubility and permeability.
The Jupyter notebook below simply calculates all available RDKit descriptors and builds a regression model. Checks that all variables were INT or FLOAT. Rows that contained NaN or very large were edited. No attempt was made to select descriptors or fine tune the model. The results were then compared to Random Forest, XGBoost and CatBoost using the same data.
The Jupyter Notebook
Clearance in Human Liver Microsomes
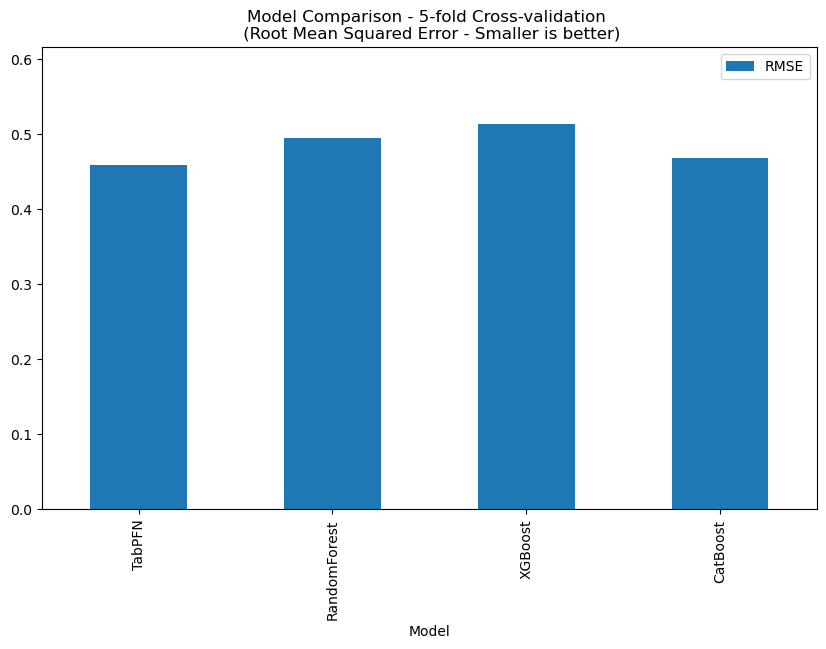
This dataset contains 3085 records, and PTabPFN RMSE: 0.4521, the comparison with other methods is shown below.
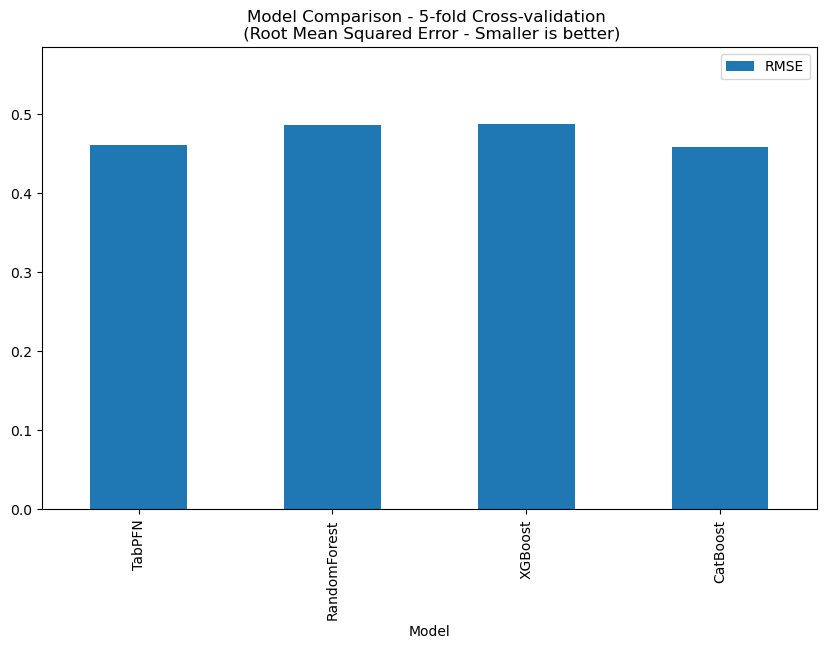
Clearance in Rat Liver Microsomes
This dataset contains 3053 records, and TabPFN RMSE: 0.5308., the comparison with other methods is shown below.
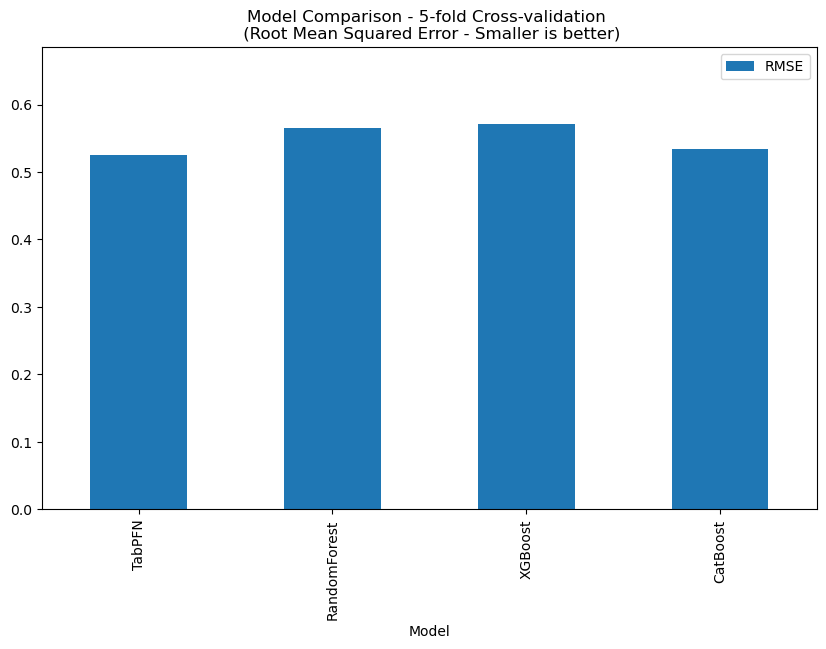
MDR1-MDCK ER (B-A/A-B)
This dataset contains 2173 records, and TabPFN RMSE: 0.411, the comparison with other methods is shown below.
Solubility
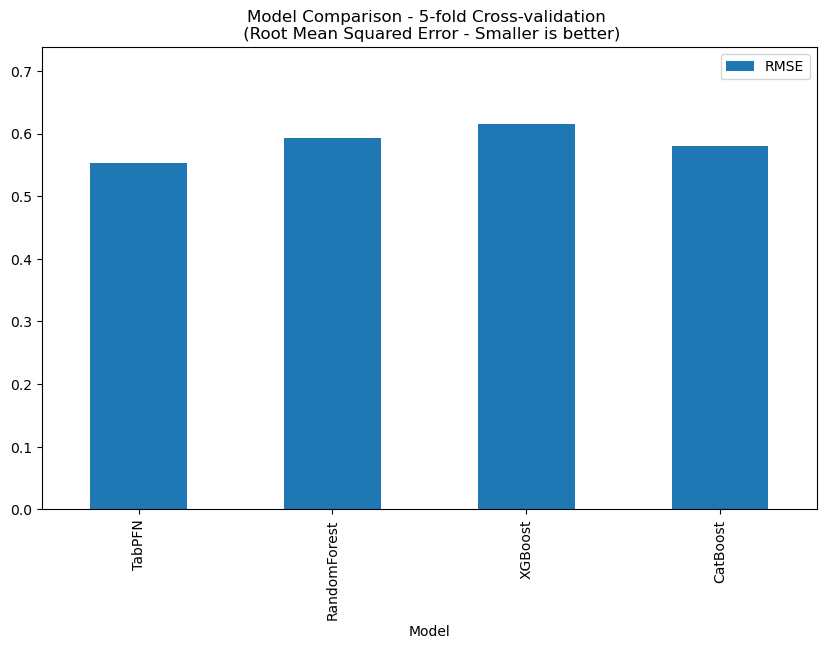
This dataset contains 2641 records, and TabPFN RMSE: 0.5240, the comparison with other methods is shown below.
Rat Plasma protein binding
This dataset contains 168 records, and TabPFN RMSE: 0.4208, the comparison with other methods is shown below.
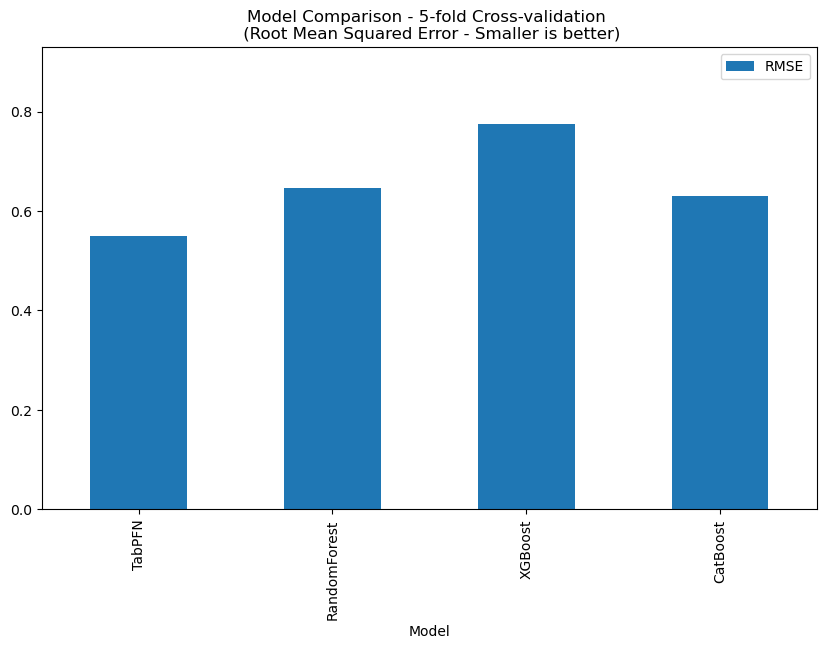
Human Plasma protein binding
This dataset contains 194 records, and TabPFN RMSE: 0.6163, the comparison with other methods is shown below.
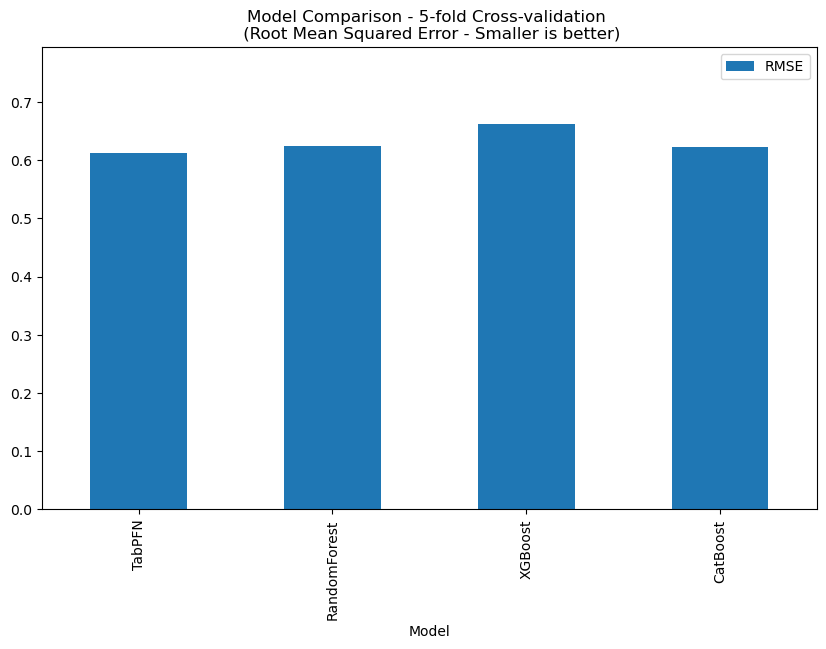
In all cases TabPFN seems to be comparable or superior to more conventional models but at this stage no detailed comparison has been undertaken.
It would also be interesting to include Chemprop and Molflux in the comparison.