When making selections from large datasets it is worth mentioning that as datasets get larger a simple random selection is often the best (and quickest) choice.
Worth reading, Relationships between Molecular Complexity, Biological Activity, and Structural Diversity http://pubs.acs.org/doi/abs/10.1021/ci0503558
None of the diversity selection methods studied, namely OptiSim, divisive K-means clustering, and self-organizing maps, yielded subsets covering the activity space of the IC50 summary data set better than subsets selected randomly
I’ve previously described a random selection script https://macinchem.org/2023/03/11/vortex-script-to-make-a-random-selection/ However, sometimes when selecting molecules for a screening collection you might want to have a few similar analogues to your randomly selected molecules to give an early indication of SAR.
This script first makes a user defined random selection, and then selects closest analogues for each molecule in the random selection.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 |
""" Select the closest compounds to each of compound from random selection. Authored by Chris (http://www.macinchem.org) """ from javax.swing import JComboBox, JPanel, JLabel, JSpinner, SpinnerNumberModel, JCheckBox import com.dotmatics.vortex.util.Layout as layout import random numsel = 10 #number to be Selected #GUI built here content = JPanel() label = JLabel("<html><b>Choose random sample</b>") content2 = JPanel() layout.fill(content2, JLabel("Number to select "), 0, 0) rangeSpinner = JSpinner(SpinnerNumberModel(numsel, 1, 100000000, 1)) layout.fill(content2, rangeSpinner, 1, 0) layout.fill(content,content2, 3, 4) #GUI displayed - when user click OK code continues ret = vortex.showInDialog(content, "Random Pick") if ret == vortex.OK: numsel = rangeSpinner.value #vortex.alert(rangeSpinner.value) #Make the random selection rows = vtable.getRealRowCount() #check if selected number is smaller than number of entries if numsel > rows: vortex.alert('Random selection larger than number of rows.') else: rowlist =[] for r in range(0, rows): vtable.setRowSelected(r, False) #remove existing selections rowlist.append(r) sublist = random.sample(rowlist, numsel) for q in range(0, len(sublist)): vtable.setRowSelected(sublist[q], True) vtable.fireVortexTableChanged(vtable.SelectionChanged) #Select number similar compounds for each cluster clusterSize = 5 #default #GUI built here content4 = JPanel() label = JLabel("<html><b>Choose cluster size</b>") content3 = JPanel() layout.fill(content3, JLabel("Size of clusters "), 0, 0) rangeSpinner = JSpinner(SpinnerNumberModel(clusterSize, 1, 100000000, 1)) layout.fill(content3, rangeSpinner, 1, 0) layout.fill(content4,content3, 3, 4) ret = vortex.showInDialog(content4, "Cluster size") if ret == vortex.OK: clusterSize = rangeSpinner.value #vortex.alert(rangeSpinner.value) structureCol = vtable.findColumnWithName("Structure") colSel = vtable.findColumnWithName("selection", True) clusterNum = "" for r in range(0, rows): response = 0 response = vtable.getRowSelected(r) if response == 1: #vortex.alert(response) clusterNum = "Cluster" + str(r) colSel.setValueFromString(r, clusterNum) simstructures = structureCol.getSimStructures(r,clusterSize) simRows = len(simstructures) for n in range(0, simRows): simRow = simstructures[n] colSel.setValueFromString(simRow, clusterNum) else: continue vtable.fireTableDataChanged() |
The first part of the script asks the user to chose the number of randomly selected molecules, these are then selected. The second dialog then asks how many should be in each cluster, and then performs a similarity search for each of the randomly selected compounds. The results are shown below in which the Cluster Number refers to the row number of the initial selected compounds in the table (which are highlighted).
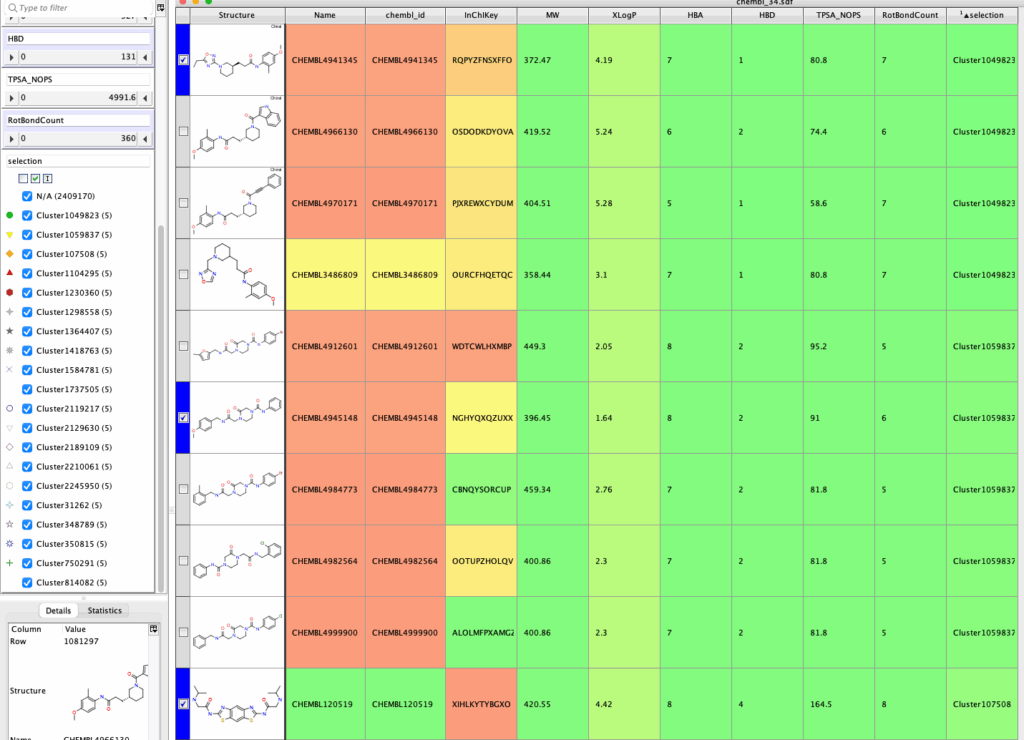
The script can be downloaded here.
