The Worldwide Protein Data Bank collects, organises and disseminates data on biological macromolecular structures. The wwPDB Partners are: PDBe, RCSB PDB, PDBj, BMRB, EMDB. Currently the PDB contains over 234,000 data files of which 73,617 are human sequences.
I’ve previously written scripts to interact with RCSB PDB, but the api has recently changed and so these needed updating. The PDBe maintains a REST API as programmatic way to obtain information from the PDB and provides comprehensive documentation.
Getting information
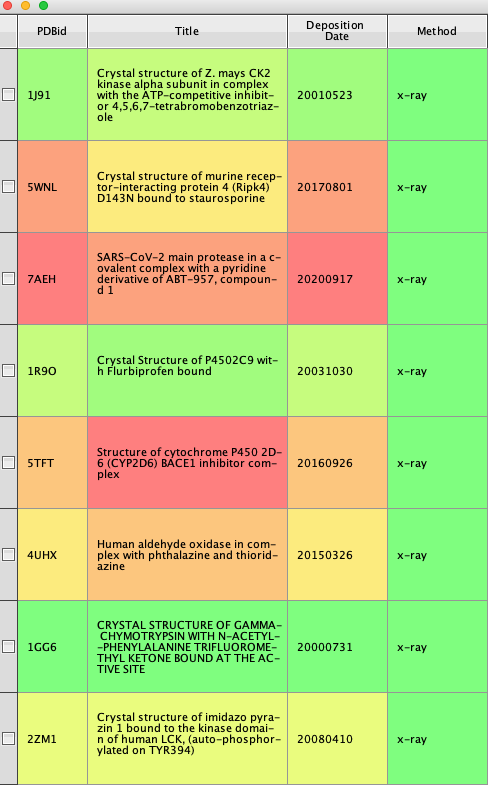
This first script takes as input a PDB ID and uses the Summary call to return a json containing a summary of properties of a PDB entry, such as the title of the entry, list of depositors, date of deposition, date of release, date of latest revision, experimental method etc.
PDBeinfo Vortex script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 |
#Use PDB id to find PDB more info #Authored by Chris Swain (http://www.macinchem.org) #Uses PDBe #https://www.ebi.ac.uk/pdbe/api/pdb/entry/summary/1cbs # Python imports import urllib2 import urllib from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os input_label = swing.JLabel("PDB column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) # Get column containing PDB id ret = vortex.showInDialog(panel, "Choose PDB column") if ret == vortex.OK: input_idx = input_cb.getSelectedIndex() if input_idx == 0: vortex.alert("you must choose a column") else: col = vtable.getColumn(input_idx - 1) #Format of query url #https://www.ebi.ac.uk/pdbe/api/pdb/entry/summary/1cbs rows = vtable.getRealRowCount() for r in range(0, int(rows)): pdbid = col.getValueAsString(r) pdbid = pdbid.lower() #needs to be lowercase #vortex.alert(pdbid) # if ":" in pdbid: #only search if pdb present # pdbid = str(pdbid)[:4] #convert to string and remove :1 api_url = "https://www.ebi.ac.uk/pdbe/api/pdb/entry/summary/" + pdbid #vortex.alert(api_url) try: molecule_record = urllib2.urlopen(api_url).read() except urllib2.HTTPError: continue j = json.loads(molecule_record) TheTitle = str(j[pdbid][0]['title']) TheDate = str(j[pdbid][0]['deposition_date']) TheMethod = str(j[pdbid][0]['experimental_method_class'][0]) coltitle = vtable.findColumnWithName('Title', 1) coltitle.setValueFromString(r, TheTitle) coldate = vtable.findColumnWithName('Deposition Date', 1) coldate.setValueFromString(r, TheDate) colmethod = vtable.findColumnWithName('Method', 1) colmethod.setValueFromString(r, TheMethod) #Data format #{"1cbs":[{"title":"CRYSTAL STRUCTURE OF CELLULAR RETINOIC-ACID-BINDING PROTEINS I AND II IN COMPLEX WITH ALL-TRANS-RETINOIC ACID AND A SYNTHETIC RETINOID","processing_site":"BNL","deposition_site":null,"deposition_date":"19940928","release_date":"19950126","revision_date":"20240207","experimental_method_class":["x-ray"],"experimental_method":["X-ray diffraction"],"split_entry":[],"related_structures":[],"entry_authors":["Kleywegt, G.J.","Bergfors, T.","Jones, T.A."],"number_of_entities":{"water":1,"polypeptide":1,"dna":0,"rna":0,"sugar":0,"ligand":1,"dna/rna":0,"other":0,"carbohydrate_polymer":0},"assemblies":[{"assembly_id":"1","name":"monomer","form":"homo","preferred":true}]}]} |
The first part of the script is simply a boilerplate containing useful Python and Vortex imports. The dialog box asks the user to select the column containing the PDB ID, this is converted to lowercase because it is in lowercase in the returned data json. The next part loops through the workspace using the pdbid to generate the api_url, urllib2 is then used to post the url and capture the response. The returned json is then parsed to select a couple of values. The new columns are generated and the appropriate data is entered. The result is shown below. The script could be modified to include extra values from the json.
Ligands Vortex script
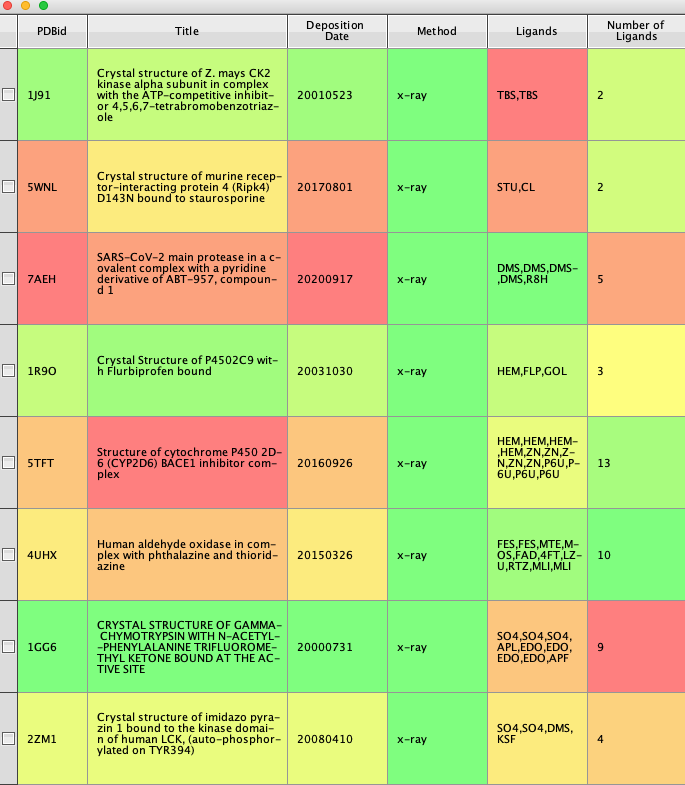
Whilst the above script gives information about the structure it does not detail the associated ligands. This script pulls the ligand information (This api call provides a a list of modelled instances of ligands, i.e. ‘bound’ molecules that are not waters.).
PDB2Ligands Vortex script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 |
#Use PDB id to find Ligand info #Authored by Chris Swain (http://www.macinchem.org) #Uses PDBe #https://www.ebi.ac.uk/pdbe/api/pdb/entry/ligand_monomers/1ls6 # Python imports import urllib2 import urllib from com.xhaus.jyson import JysonCodec as json # Vortex imports import com.dotmatics.vortex.util.Util as Util import com.dotmatics.vortex.mol2img.jni.genImage as genImage import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img import jarray import binascii import string import os input_label = swing.JLabel("PDB column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) # Get column containing PDB id ret = vortex.showInDialog(panel, "Choose PDB column") if ret == vortex.OK: input_idx = input_cb.getSelectedIndex() if input_idx == 0: vortex.alert("you must choose a column") else: col = vtable.getColumn(input_idx - 1) #Format of query url #https://www.ebi.ac.uk/pdbe/api/pdb/entry/ligand_monomers/1ls6 rows = vtable.getRealRowCount() for r in range(0, int(rows)): pdbid = col.getValueAsString(r) pdbid = pdbid.lower() #needs to be lowercase #vortex.alert(pdbid) # if ":" in pdbid: #only search if pdb present # pdbid = str(pdbid)[:4] #convert to string and remove :1 api_url = "https://www.ebi.ac.uk/pdbe/api/pdb/entry/ligand_monomers/" + pdbid #vortex.alert(api_url) try: molecule_record = urllib2.urlopen(api_url).read() except urllib2.HTTPError: continue j = json.loads(molecule_record) ThenumChemID = len(j[pdbid]) ChemIDs = [] for i in range(ThenumChemID): TheChemID = str(j[pdbid][i]["chem_comp_id"]) ChemIDs.append(TheChemID) ChemIDs = ",".join(ChemIDs) #vortex.alert(ChemIDs) colchemId= vtable.findColumnWithName("Ligands", 1) colchemId.setValueFromString(r, ChemIDs) colnumchemId= vtable.findColumnWithName("Number of Ligands", 1) colnumchemId.setValueFromString(r, str(ThenumChemID)) vtable.fireTableStructureChanged() #Data Format #{"1ls6":[{"chain_id":"A","author_residue_number":2001,"author_insertion_code":"","chem_comp_id":"A3P","alternate_conformers":0,"entity_id":2,"struct_asym_id":"B","residue_number":1,"chem_comp_name":"ADENOSINE-3'-5'-DIPHOSPHATE","weight":427.201,"carbohydrate_polymer":false,"branch_name":""},{"chain_id":"A","author_residue_number":3001,"author_insertion_code":"","chem_comp_id":"NPO","alternate_conformers":0,"entity_id":3,"struct_asym_id":"C","residue_number":1,"chem_comp_name":"P-NITROPHENOL","weight":139.109,"carbohydrate_polymer":false,"branch_name":""},{"chain_id":"A","author_residue_number":4001,"author_insertion_code":"","chem_comp_id":"NPO","alternate_conformers":0,"entity_id":3,"struct_asym_id":"D","residue_number":1,"chem_comp_name":"P-NITROPHENOL","weight":139.109,"carbohydrate_polymer":false,"branch_name":""}]} |
The majority of the script is similar to the previous script, in this case however there are likely to be multiple ligands so we loop through the json and then convert the resulting list of ligand ID to a string for inserting into the Vortex workspace. The result is shown below
The two vortex scripts can be downloaded here.
