Drug-induced liver injury (DILI) is a significant concern in drug discovery, in particular because it might only be observed late in large clinical studies, after significant costs and resources have been invested. Any way to flag potential issues early in the drug discovery process is to be welcomed, allowing early investigation of potential toxicity.
Dilipred DOI is an interesting in-silico tool that uses a variety of different approaches and data sources to identify potential for hepatotoxicity.
All code is available on GitHub https://github.com/srijitseal/DILI/tree/main and can be installed using pip.
|
1 |
pip install dilipred |
Since dilipred requires Python <3.12, >=3.9 I actually created a conda environment with python 3.10 and then installed dilipred within that environment.
It can then be accessed via the command line
|
1 |
% dilipred -smiles "CC1=C(C2=CC(N3CCN(C)CC3)=NC=C2N(C(C(C)(C4=CC(C(F)(F)F)=CC(C(F)(F)F)=C4)C)=O)C)C=CC=C1" |
The results being saved in a time stamped csv file. Whilst this works is does mean that if you run a number of different molecules then remembering which is which based on a timestamp file name is not always ideal. I got in touch with the authors to see if they could add an option for the output be directed to a specific named file. Within 24 hours dilipred had been updated!
|
1 |
pip install dilipred --upgrade |
A quick update using pip and the new version was accessible, and the output was now directed to a named file.
|
1 |
% dilipred -smiles "CC1=C(C2=CC(N3CCN(C)CC3)=NC=C2N(C(C(C)(C4=CC(C(F)(F)F)=CC(C(F)(F)F)=C4)C)=O)C)C=CC=C1" --output "myfile" |
The results look like this, and the .csv file can be opened by a variety of applications.
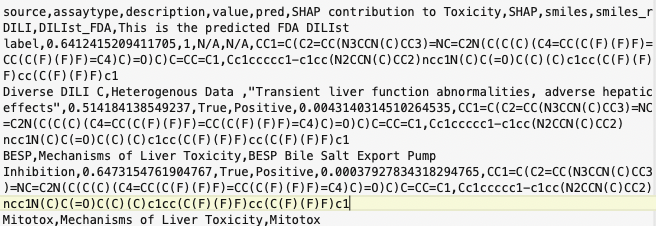
Accessing DILIpred from Vortex
As well as accessing scripts from the “Scripts” menu right-click on a structure brings a contextual menu that allows the user to select scripts from the context folder within the Vortex_Add-ons folder.
The Vortex script below generates a SMILES string from the structure in the workspace, then asks the user to select an identifier that will be used as the filename. This file will be saved in a temporary directory (you can edit this to a particular directory if you want to keep the .csv files). The command is then created and run. The output file is then read by Vortex to create a new workspace containing all the results.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 |
#Drug induced Liver injury prediction #Improved Detection of Drug-Induced Liver Injury by Integrating Predicted In Vivo and In Vitro Data #Srijit Seal, Dominic Williams, Layla Hosseini-Gerami, Manas Mahale, Anne E. Carpenter, Ola Spjuth, #and Andreas Bender #doi: https://doi.org/10.1021/acs.chemrestox.4c00015 import sys import com.dotmatics.vortex.util.Util as Util import subprocess import csv import tempfile smiles = '' col = vtable.findColumnWithName('Structure', 0) if (col == None): col = vtable.findColumnWithName('SMILES', 0) if (col == None): vortex.alert('Load a workspace with a Structure or SMILES column please.') quit() else: inputsmiles = col.getValueAsString(cell_row) else: inputsmiles = vortex.getMolProperty(vtable.getMolFileManager().getMolFileAtRow(cell_row), 'SMILES') inputsmiles = inputsmiles.encode('utf-8') #for testing #vortex.alert('SMILES = '+inputsmiles) #Need an identifier for file name input_label = swing.JLabel("ID column (for input)") input_cb = workspace.getColumnComboBox() panel = swing.JPanel() layout.fill(panel, input_label, 0, 0) layout.fill(panel, input_cb, 1, 0) # Get name of column containing the compound ID ret = vortex.showInDialog(panel, "Choose CID column") input_idx = input_cb.getSelectedIndex() col = vtable.getColumn(input_idx - 1) if (col == None): vortex.alert('Load a workspace with a parent_cmpd_chemblid column please.') quit() else: taskID = col.getValueAsString(cell_row) #need to save dilipred output tmpdir = tempfile.mkdtemp() #for testing #outputfile = '/Users/username/Public/'+ taskID + '.csv' outputfile = tmpdir + "/" + taskID + '.csv' #Demo command #dilipred -smiles "CC1=C(C2=CC(N3CCN(C)CC3)=NC=C2N(C(C(C)(C4=CC(C(F)(F)F)=CC(C(F)(F)F)=C4)C)=O)C)C=CC=C1" --output "myfile" #Need path to dilipred subprocess.call(['/Users/chrisswain/miniconda3/bin/dilipred', '-smiles', inputsmiles, '--output', outputfile]) import com.dotmatics.vortex.table.VortexTableModel as vtm vortex.loadAnyFile(outputfile) vtable.fireTableStructureChanged() |
Right-click on the structure to select the script.
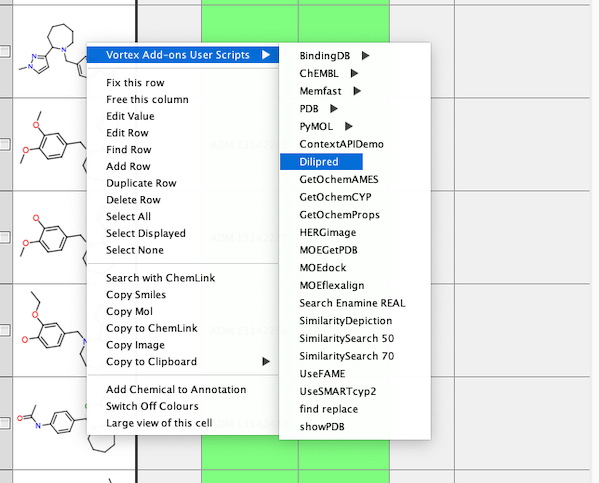
Then choose the identifier

The calculation takes a few seconds and will be displayed in a new workspace.
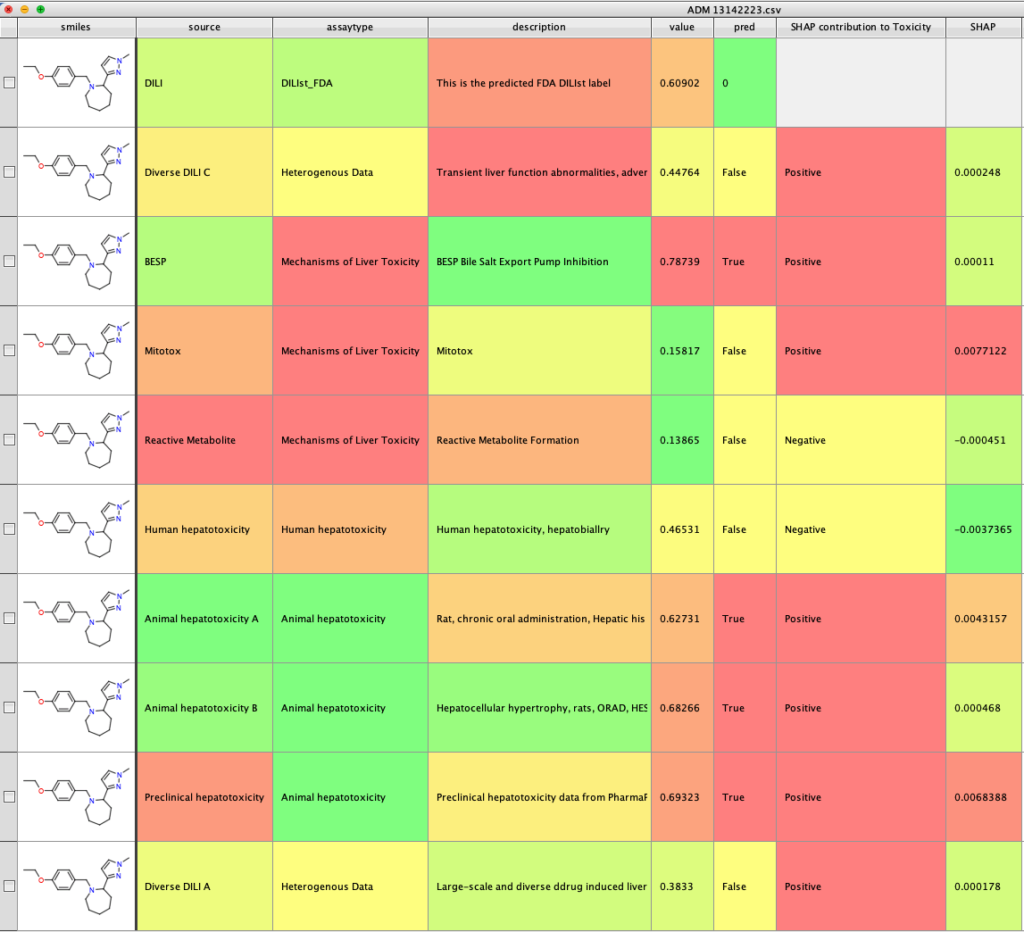
This script illustrates the process for accessing a command line program to run an in-silico prediction and display the results. The python script could be modified for other applications.
Improved Detection of Drug-Induced Liver Injury by Integrating Predicted In Vivo and In Vitro Data
Srijit Seal, Dominic Williams, Layla Hosseini-Gerami, Manas Mahale, Anne E. Carpenter, Ola Spjuth, and Andreas Bender
doi: https://doi.org/10.1021/acs.chemrestox.4c00015

