Augmented reality is finding new applications in science, in particular the ability to enhance publications or lecture notes, and viewers can set up a free account with [Augment](http://www.augment.com/) to provide easy access.
A number of applications can be used to create the files required for 3D display
* Jmol application (not applet) export as VRML (.wrl format) or Wavefront (.obj format)<br>
* PYMOL export as VRML2 (.wrl format).<br>
* VMD export as VRML (.wrl format) or .stl format.<br>
* MOE using io_vrml.svl script export as VRML (.wrl format)<br>
* UCSF Chimera can export crystal structures as STL and VRML files<br>
All work fine but can require command line access to generate the required files and some file manipulation. There is a tutorial here.
I was asked recently if it might be possible to generate an AppleScript droplet that you could simply drop a chemical structure file onto to generate the desired files needed for the Augment, and this is an ideal use case for a droplet.
Droplets are specialised AppleScript applets which are designed to process items dragged onto them. Their icon contains a downward pointing arrow indicating their status as a droplet.
This script uses Jmol to generate the Wavefront .obj and .mtl files. Jmol can be downloaded from sourceforge https://sourceforge.net/projects/jmol/files/Jmol/ and downloads to yield a folder named in this format Jmol-xx.xx.xx. This is a bit of a pain because the script requires the exact path to the executable.
Jmol is a free, open source molecule viewer for students, educators, and researchers in chemistry, biochemistry, physics, and materials science. It is cross-platform, running on Windows, Mac OS X, and Linux/Unix systems
The JSmol JmolApplet is a web browser JavaScript application that can be integrated easily into web pages. Though Java-based for development and management, JSmol is completely JavaScript.
The Jmol application is a stand-alone Java application (Jmol.jar) that runs on the desktop. It has all the capabilities of the JSmol applet but runs approximately 3-6 times faster. In addition, it can be run in “headless” mode (JmolData.jar) in order to carry out tasks that need to be replicated. Jmol.jar can be integrated into any other Java program, providing file loading, structure processing, viewing, and output capabilities.
The command to create the files needed.
java -jar /Applications/jmol-14.29.17/JmolData.jar -j write OBJ /Users/username/Desktop/SampleFiles/carbocation.obj /Users/username/Desktop/SampleFiles/carbocation.xyz
This actually creates two files /carbocation.obj and /carbocation.mtl which is the material file that defines the surface
Whilst you can open and view these files in an application like Meshlab you need to compress these two files into a single Archive.zip file for uploading to the Augment site. We can use another command line application to generate the zip file.
man zip
ZIP(1L)
NAME
zip – package and compress (archive) files
SYNOPSIS
zip [-aABcdDeEfFghjklLmoqrRSTuvVwXyz!@$] [–longoption …] [-b path] [-n suffixes] [-t date] [-tt date] [zipfile [file …]] [-xi list]
The AppleScript
Droplets are written in a specific manner using the on open handler. This droplet will process only files which have been dragged onto its icon. It will not process folders, alias files, or files within folders.
The first step gets the list of files that have been dropped onto the icon, we then get all the file information for each file, you could use this to limit the number of filetypes that can be processed. We then need to generate the paths to the source file and the output files, in this script the output files are created in the same folder as the input files. These paths will be Mac paths so we need to convert Mac file paths (colon delimited) to unix paths (slash delimited).
set unix_path to quoted form of POSIX path of new_path
We can now construct the Jmol command, remember you need to include the full path to the JmolData.jar, in this case the Jmol folder is in the Applications folder, if you have installed it elsewhere or have a different version this will need to be updated.
set myscript to "java -jar /Applications/jmol-14.29.17/JmolData.jar -j 'write OBJ " & unix_path & "' " & input_file
The next step is to use the do shell script to run the Jmol command.
This will create the .obj and .mtl files, we next use the zip command to compress the two files into a single archive.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 |
on open source_files repeat with i from 1 to number of items in source_files set source_file to item 1 of source_files --You can comment out the dialogs if needed --display dialog source_file as text set input_file to POSIX path of source_file --display dialog input_file set file_name to name of (info for alias source_file) set file_extension to name extension of (info for alias source_file) set file_type to file type of (info for alias source_file) set file_kind to kind of (info for alias source_file) --display dialog file_name --display dialog file_extension --display dialog file_type --display dialog file_kind set the trimmed_name to text 1 thru -((length of file_extension) + 2) of the file_name --display dialog trimmed_name set the trimmed_path to text 1 thru -((length of file _extension) + 2) of the (source_file as text) set new_name to trimmed_name & ".obj" set new_path to trimmed_path & ".obj" set new_path2 to trimmed_path & ".mtl" --this path is where the new file appears in the same folder as the dropped file was located --command to generate file --You may need to modify path to Jmol and version set myscript to "java -jar /Applications/jmol-14.29/JmolData.jat -j 'write OBJ " & unixpath & "' " & input_file --diaplay dialog myscript do shell script myscript --now compress to zip file set zippath to trimmed_path & ".zip" set unixzip to POSIX path of zippath set zipscript to "zip -r " & unixzip & " " & unix_path & " " & unix_path2 do shell script zipscript end repeat end open |
If you drag and drop chemical structure files onto the droplet you should end up with results like this, with a new .obj, .mtl and .zip file created for each of the structure files. Jmol can read a variety of different chemistry file formats described here with examples https://sourceforge.net/p/jmol/code/HEAD/tree/trunk/Jmol-datafiles/
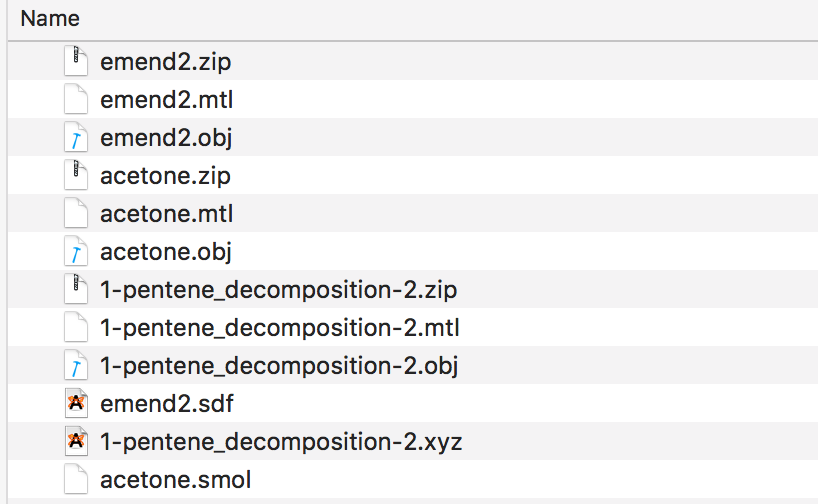
The instructions on how to upload to Augment are available here.
You can download the droplet here
Nick Greeves has tweeted an example of its use and a demo page here
Last Updated 9 August 2018
