Sometimes I have two datasets and I just want to know the overlap of identical structures. This script counts the number of identical structures by comparing InChIKeys. We start by reading two files into separate workspaces.
he next part of the script generates the InChiKey for each molecule in both workspaces. We then check for duplicates first in each table, and then for duplicates between the tables. A new workspace is then generated with the results as shown below.
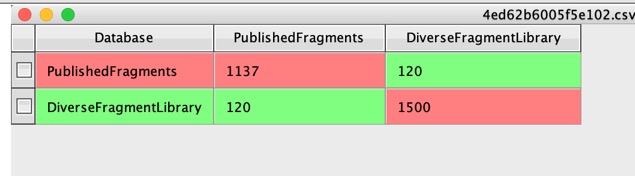
The figure in the top left of the Matrix (1137) is the number of unique structures there are in “PublishedFragments”, the number in the bottom right (1500) is the number of unique structures in DiverseFragmentLibrary. In the case of “PublishedFragments” this is actually less than the numbers in the workspace, this is because there are a number of duplicate structures in that file. The figure of 120 corresponds to how many identical structures there are between the two datasets.
The Vortex Script
# A script to count identical structures in two workspaces
# Vortex imports
import com.dotmatics.vortex.util.Util as Util
import com.dotmatics.vortex.mol2img.jni.genImage as genImage
import com.dotmatics.vortex.mol2img.Mol2Img as mol2Img
import jarray
import binascii
import string
import os
import sys
from collections import defaultdict
mycount = vortex.getWorkspaceCount() #number of workspaces
col_names = ["Database"] #array
results = [] #2D array
inchikeys1 = defaultdict(list)
vws = vortex.getWorkspace(0) #first workspace
vtable1 = vws.getTable()
myname1 = vws.getName()
#vortex.alert(myname1)
col_names.append(myname1)
InChIKeyColumn = vtable1.findColumnWithName("InChIKey",1)
rows = vtable1.getRealRowCount()
for r in range(0, int(rows)):
try:
mol = vtable1.molFileManager.getMolFileAtRow(r)
inChIKey = vortex.getMolProperty(mol, 'InChIKey')
InChIKeyColumn.setValueFromString(r, inChIKey)
inchikeys1[inChIKey].append(r)
except:
pass
vtable.fireTableStructureChanged()
inchikeys2 = defaultdict(list)
vws = vortex.getWorkspace(mycount -1) #second workspace
vtable2 = vws.getTable()
myname2 = vws.getName()
InChIKeyColumn = vtable2.findColumnWithName("InChIKey",1)
rows = vtable2.getRealRowCount()
for r in range(0, int(rows)):
try:
mol = vtable2.molFileManager.getMolFileAtRow(r)
inChIKey = vortex.getMolProperty(mol, 'InChIKey')
InChIKeyColumn.setValueFromString(r, inChIKey)
inchikeys2[inChIKey].append(r)
except:
pass
vtable.fireTableStructureChanged()
#looking for duplicates within each table
dupsdict1 = [k for k in inchikeys1 if k in inchikeys1]
len_dupsdict1 = len(dupsdict1)
dupsdict2 = [k for k in inchikeys2 if k in inchikeys2]
len_dupsdict2 = len(dupsdict2)
#looking fr duplicates between tables
dups = [k for k in inchikeys1 if k in inchikeys2]
len_dups = len(dups)
vortex.alert(len_dups)
row1 = [myname1, len_dupsdict1 , len_dups]
row2 = [myname2, len_dups, len_dupsdict2]
col_names.append(myname2)
results.append(row1)
results.append([])
results.append(row2)
#create new workspace with the results
arrayToWorkspace(results, col_names, 'Identical')
The script can be downloaded here
Last updated 7 March 2018
