A recent publication described BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification DOI. There are a number of tools that predict sites of metabolism on a molecule and I’ve mentioned a couple FAME and SMARTCyp in the past. These packages flag potential metabolic hot spots (mainly for CYP mediated metabolism) but don’t attempt to provide any information on the putative metabolites.
BioTransformer combines a machine learning approach with a knowledge-based approach to predict small molecule metabolism in human tissues (e.g. liver tissue), the human gut as well as the environment (soil and water microbiota), via its metabolism prediction tool. In addition BioTransformer provides information on secondary metabolism.
A comprehensive evaluation of BioTransformer showed that it was able to outperform two state-of-the-art commercially available tools (Meteor Nexus and ADMET Predictor), with precision and recall values up to 7 times better than those obtained for Meteor Nexus or ADMET Predictor on the same sets of pharmaceuticals, pesticides, phytochemicals or endobiotics under similar or identical constraints.
BioTransformer is available as an open access command-line tool (GNU GENERAL PUBLIC LICENSE Version 2), or a software library. It is freely available at https://bitbucket.org/djoumbou/biotransformerjar/. If you are not comfortable using the command line it can be accessed via an open access RESTful application at www.biotransformer.ca where you can simply paste in a SMILES string or upload an sdf file.
BioTransformer consists of a metabolism prediction tool (BMPT), and a metabolite identification tool (BMIT). The BMPT consists of five independent prediction modules called “transformers”, namely: (1) the Enzyme Commission based (EC-based) transformer, (2) the CYP450 (phase I) transformer, (3) the phase II transformer, (4) the human gut microbial transformer, and (5) the environmental microbial transformer. For the prediction of metabolites, BioTransformer implements two approaches, a rule-based or knowledge-based approach, and a machine learning approach. BioTransformer’s knowledge-based system consists of three major components: (1) a biotransformation database (called MetXBioDB) containing detailed annotations of experimentally confirmed metabolic reactions, (2) a reaction knowledgebase containing generic biotransformation rules, preference rules, and other constraints for metabolism prediction, and (3) a reasoning engine that implements both generic and transformer-specific algorithms for metabolite prediction and selection. The BMPT machine learning system uses a set of random forest and ensemble prediction models for the prediction of CYP450 substrate selectivity, and for the Phase II filtering of molecules. BioTransformer’s Metabolite Identification Tool builds on the BMPT to identify specific metabolites using mass spectrometry (MS) data, namely accurate mass or chemical formula information.
I downloaded it and it is perhaps worth noting that you also need to download the folders database and support files, and save them in the same folder as the .jar file.
The help can be called from the command line
(base) MacPro:biotransformer username$ java -jar biotransformer-1-0-8.jar -h
usage:
java -jar biotransformer-1.0.8 [-a] -b <BioTransformer Option> [-f
<Formulas>] [-h] [-imol <MOL Input>] [-isdf <Sdf Input>] [-ismi
<SMILES Input>] -k <BioTransformer Task> [-m <Masses>] [-ocsv <Csv
Output>] [-osdf <Sdf Output>] [-s <Number of steps>] [-t <Mass
Tolerance>]
This is the version 1.0.8 of BioTransformer. BioTransformer is a software tool that predicts small molecule metabolism in mammals, their gut microbiota, as well as the soil/aquatic microbiota. BioTransformer also assists scientists in metabolite identification, based on the metabolism prediction.
-a,--annotate Search PuChem for each product, and
store with CID and synonyms, when
available.
-b,--btType <BioTransformer Option> The type of description: Type of
biotransformer - EC-based
(ecbased), CYP450 (cyp450), Phase
II (phaseII), Human gut microbial
(hgut), human super transformer*
(superbio, or allHuman),
Environmental microbial
(envimicro)**.
If option -m is enabled, the only
valid biotransformer types are
allHuman, superbio and env.
-f,--formulas <Formulas> Semicolon-separated list of
formulas of compounds to identify
-h,--help Prints the usage.
-imol,--molinput <MOL Input> The input, which can be a Mol file
-isdf,--sdfinput <Sdf Input> The input, which can be an SDF
file.
-ismi,--ismiles <SMILES Input> The input, which can be a SMILES
string
-k,--task <BioTransformer Task> The task to be permed: pred for
prediction, or cid for compound
identification
-m,--masses <Masses> Semicolon-separated list of masses
of compounds to identify
-ocsv,--csvoutput <Csv Output> Select this option to return CSV
output(s). You must enter an output
filename
-osdf,--sdfoutput <Sdf Output> Select this option to return SDF
output(s). You must enter an output
filename
-s,--nsteps <Number of steps> The number of steps for the
prediction. This option can be set
by the user for the EC-based,
CYP450, Phase II, and Environmental
microbial biotransformers. The
default value is 1.
-t,--mTolerance <Mass Tolerance> Mass tolerance for metabolite
identification (default is 0.01).
In addition the help also provides a few examples and tips
To predict the 2-step biotransformation of Thymol (a monoterpene) using the human super transformer (option allHuman) using the SMILES input and saving to a CSV file, run
java -jar biotransformer-1-0-8.jar -k pred -b allHuman -ismi "CC(C)C1=CC=C(C)C=C1O" -ocsv #{replace with output file name} -s 2
DO NOT forget the quotes around the SMILES string or the list of masses,
Running the application
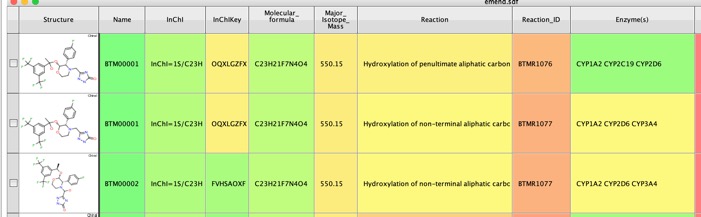
I tried this out with the structure of Emend which I also used when looking at FAME and SMARTCyp.
(base) MacPro:biotransformer username$ java -jar biotransformer-1-0-8.jar -k pred -b allHuman -ismi "FC(F)(F)c1cc(cc(c1)C(F)(F)F)[C@H](O[C@H]4OCCN(CC/2=N/C(=O)NN\2[C@H]4c3ccc(F)cc3)C" -osdf /Users/username/Projects/biotransformer/emend.sdf -s 2
…..
Step: 1
Step: 2
Biotransformations: 235
Unique Biotransformations: 235
The whole process took around 90 seconds and the results were written to an sdf file and were displayed in Vortex. As well as the putative metabolite structure a range of other properties are calculated including InChiKey which can be used to search many other databases, Major Isotopic Mass which could be used in metabolite identification. Together with a description of the reaction and the likely enzymes involved. For the prediction of CYP450 metabolism, BioTransformer makes use of CypReact, a tool for CYP450 substrate specificity prediction.
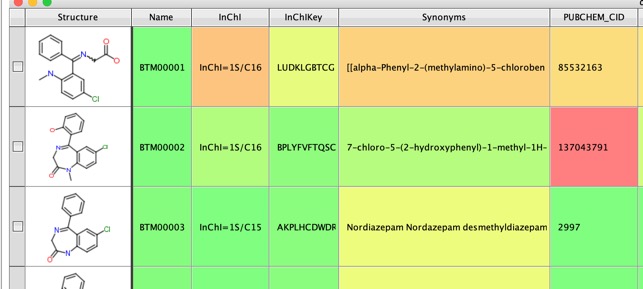
If we use Diazepam as the query molecule and add the -a option to search PubChem for each product, and annotate with CID and synonyms, when available.
java -jar biotransformer-1-0-8.jar -k pred -b allHuman -ismi "CN1C2=C(C(C3=CC=CC=C3)=NCC1=O)C=C(Cl)C=C2" -osdf /Users/username/Projects/biotransformer/diazepam.sdf -s 2 -a
The results now include synonyms and the Pubchem CID which can be searched for further information on the structure.
Running the process of Diazepam yielded a sdf file containing yielded 151 structures but on closer examination there were many duplicate structures and only 51 unique structures (I used this Vortex script to flag duplicate structures). This is because several reaction patterns can yield the same structure. Around 30 had PubChem CID suggesting these are previously identified metabolites.
Looking at the results in more detail
One thing that is readily obvious is that BioTransformer is very comprehensive, this means that it suggests potential metabolites that you may never actually see in vivo, it also does not give any information about the rate at which the metabolites might be formed or which are the most likely. This is absolutely invaluable for anyone involved in metabolite identification but might be a little daunting for a medicinal chemist simply wanting to know which site to address to block metabolism, however as I mentioned the total number of putative metabolites will include duplicate structures and the number of unique structures will be much fewer.
To further aid metabolite ID BioTransformer calculates metabolite major isotopic weight but also provides the precursor major isotopic weight and precursor InChiKey, I can see that BioTransformer could easily incorporated into a metabolite ID workflow and be used to elucidate metabolic pathways.
Last Updated 4 May 2019