Here are two variations of a Jupyter Notebook to help with docking experiments. The first version runs locally and requires the user to install RDKit, OpenBabel, SMINA and py3Dmol, the second version can be run using Google CoLab and thus all you require is a web browser.
This notebook implements a typical protocol for docking ligands to a target protein. It uses RDKit (http://www.rdkit.org) to generate a number of reasonable conformations for each ligand and then uses SMINA (https://sourceforge.net/projects/smina/) to do the docking. Two methods of docking are implemented, the first docks into a rigid receptor, the second optional approach sets the protein side-chains around the active site to be flexible. Bear in mind flexible docking will be much, much slower. In the next step the resulting docked poses are rescored using a random forest model described in this publication DOI. Finally a selection of the docked structures are viewed using py3Dmol.
You can view the complete notebook here and you can download a folder containing the notebook and the necessary files here.
The downloaded folder contains two notebooks, ConformationGenerationDocking.ipynb is the one for local use,
You will need to install a variety of packages using conda
conda config --add channels conda-forge
conda install jupyter
conda install -c rdkit rdkit
conda install numpy
conda install scipy
conda install scikit-learn
conda install pandas
conda install matplotlib
conda install seaborn
conda install openbabel
conda install py3dmolSMINA is a command-line application for docking. Instructions for installation are on the website.
Rescore using a random forest model described in https://doi.org/10.1038/srep46710
Download from GitHub
Once installed you can start the notebook, the other files you need to run the notebook are in the downloaded folder these include the protein for docking (proteinminusligand.pdb) and a small selection of compounds for docking (asinexSelectionexport.sdf).
Using GoogleCoLab
RSC CICAG has hosted a number of workshops for key Open-Source chem software, these workshops are now all available on YouTube. In particular there is an excellent introduction to using GoogleCoLab from Jan Jensen.
If you want to use the notebook without the need to install anything one option is to use GoogleCoLab.
Colaboratory, or “Colab” for short, allows you to write and execute Python in your browser, with Zero configuration required, Free access to GPUs, Easy sharing
In a web browser go to https://colab.research.google.com/notebooks/intro.ipynb and there click on the “File” menu to upload the notebook (DockingCoLab.ipynb).

Then click on the upload icon (highlighted in red) to upload the rest of the files, you may need to click on the orange folder icon.
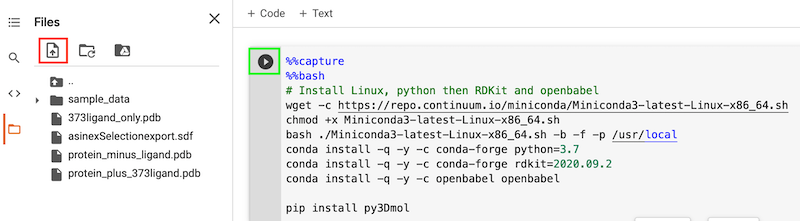
You can now click on the arrow on each cell to run it. The first cell installs Linux, Python and some of the packages needed (note we need Linux versions).
The more observant will have spotted a specific version of RDKIt is being used.
For some reason core libs are being loaded from the system packages in the colab environment. Until now, by luck, they matched the version provided by conda-forge, so everything worked. However, conda-forge recently updated to a newer version of compilers, and they got used in the RDKit 2020.09.3 release. So easiest fix for now is to pin exactly rdkit 2020.09.2. But better fix is to figure out how to ensure the libs are loaded from conda forge (they are installed correctly, just seem to be lower priority when loading…)
Of course one of the really cool things about CoLabs is the ability to share notebooks and to have several people working on them at the same time. Just click on the “Share” button and add email addresses and get a shareable link. It is rather wonderful to be able to sit there and watching code appear magically in a cell.
Many thanks to Matt and David for their “collaborative” invaluable help.
Last Updated 14 December 2020.