https://www.rdkit.org
The RDKit is an open source toolkit for cheminformatics, 2D and 3D molecular operations, descriptor generation for machine learning, etc. There’s also a molecular database cartridge for PostgreSQL and cheminformatics nodes for KNIME (distributed from the KNIME community site: https://www.knime.org/rdkit)
The RDKit core algorithms and data structures are written in C++. Wrappers are provided to use the toolkit from either Python (2.x and 3.x), Java, or C#.
RDKIt was installed on both machines using miniconda
|
1 |
conda install -c rdkit |
There are a standard set of benchmarks that run with the RDKit in order to detect systematic performance improvements or regressions. Those are here:
https://github.com/rdkit/rdkit/blob/master/Regress/Scripts/new_timings.py
https://github.com/rdkit/rdkit/blob/master/Regress/Scripts/new_timings.py
The associated data files are in the folder
https://github.com/rdkit/rdkit/blob/master/Regress/Data
The scripts run through a variety of cheminformatics operations
The command used was
|
1 |
python new_timings.py |
The script was run 3 times and the fastest times shown below.
The Results
| Intel time | M1 max time | M2 Air time | M2 Mac Studio Ultra |
|---|---|---|---|
| INFO: mols from smiles Results1: 11.08 seconds, 50000 passed, 0 failed INFO: Writing: Canonical SMILES Results2: 4.99 seconds INFO: mols from sdf Results1: 3.96 seconds, 10000 passed, 0 failed INFO: patterns from smiles Results3: 0.04 seconds, 823 passed, 0 failed INFO: Matching1: HasSubstructMatch Results4: 22.94 seconds INFO: Matching2: GetSubstructMatches Results5: 22.94 seconds INFO: reading SMARTS Results6: 0.01 seconds for 428 patterns INFO: Matching3: HasSubstructMatch Results7: 90.56 seconds INFO: Matching4: GetSubstructMatches Results8: 85.82 seconds INFO: Writing: Mol blocks Results10: 15.20 seconds INFO: BRICS decomposition Results11: 27.28 seconds INFO: Generate 2D coords Results12: 9.39 seconds INFO: Generate topological fingerprints Results16: 78.53 seconds INFO: Generate morgan fingerprints Results16: 3.31 second | INFO: mols from smiles Results1: 4.11 seconds, 50000 passed, 0 failed INFO: Writing: Canonical SMILES Results2: 2.16 seconds INFO: mols from sdf Results1: 1.60 seconds, 10000 passed, 0 failed INFO: patterns from smiles Results3: 0.02 seconds, 823 passed, 0 failed INFO: Matching1: HasSubstructMatch Results4: 13.55 seconds INFO: Matching2: GetSubstructMatches Results5: 13.67 seconds INFO: reading SMARTS Results6: 0.01 seconds for 428 patterns INFO: Matching3: HasSubstructMatch Results7: 56.01 seconds INFO: Matching4: GetSubstructMatches Results8: 50.21 seconds INFO: Writing: Mol blocks Results10: 7.35 seconds INFO: BRICS decomposition Results11: 13.51 seconds INFO: Generate 2D coords Results12: 4.88 seconds INFO: Generate topological fingerprints Results16: 51.80 seconds INFO: Generate morgan fingerprints Results16: 1.61 seconds | INFO: mols from smiles Results1: 3.9 seconds, 50000 passed, 0 failed INFO: Writing: Canonical SMILES Results2: 2.36 seconds INFO: mols from sdf Results1: 1.64 seconds, 10000 passed, 0 failed INFO: patterns from smiles Results3: 0.02 seconds, 823 passed, 0 failed INFO: Matching1: HasSubstructMatch Results4: 13.13 seconds INFO: Matching2: GetSubstructMatches Results5: 12.96 seconds INFO: reading SMARTS Results6: 0.01 seconds for 428 patterns INFO: Matching3: HasSubstructMatch Results7: 50.4seconds INFO: Matching4: GetSubstructMatches Results8: 45.82 seconds INFO: Writing: Mol blocks Results10: 6.83 seconds INFO: BRICS decomposition Results11: 17.51 seconds INFO: Generate 2D coords Results12: 4.65 seconds INFO: Generate topological fingerprints Results16: 46.80 seconds INFO: Generate morgan fingerprints Results16: 1.36 seconds | INFO: mols from smiles Results1: 3.3 seconds, 50000 passed, 0 failed INFO: Writing: Canonical SMILES Results2: 1.9 seconds INFO: mols from sdf Results1: 1.3 seconds, 10000 passed, 0 failed INFO: patterns from smiles Results3: 0.01 seconds, 823 passed, 0 failed INFO: Matching1: HasSubstructMatch Results4: 13.13 seconds INFO: Matching2: GetSubstructMatches Results5: 13.0 seconds INFO: reading SMARTS Results6: 0.01 seconds for 428 patterns INFO: Matching3: HasSubstructMatch Results7: 52 seconds INFO: Matching4: GetSubstructMatches Results8: 47 seconds INFO: Writing: Mol blocks Results10: 6.8 seconds INFO: BRICS decomposition Results11: 17.51 seconds INFO: Generate 2D coords Results12: 4.3 seconds INFO: Generate topological fingerprints Results16: 38.9 seconds INFO: Generate morgan fingerprints Results16: 1.2 seconds |
These all measure single core performance.
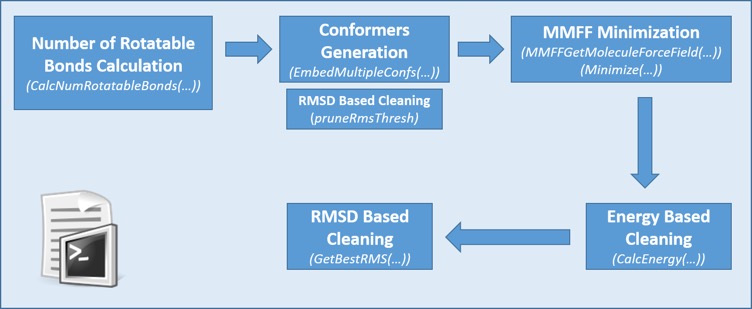
Pharmacelera have created an open-source python script for conformation generation genConf.py. This script generates conformations plus a number of filters to generate a diverse selection of reasonable conformations. This is very typical workflow and as such is good measure of likely performance benefit.
genConf.py script workflow generated by Pharmacelera
The script is available for download here
Link to conformer script 3.0: https://pharmacelera.com/rdkit-conformer-generation-script-python-3/
Link to conformer script 2.7: https://pharmacelera.com/blog/scripts/rdkit-conformation-generation-script/
Again I used a selection of 1000 random structures from ChEMBL.
The Intel MacBook Pro took 4 hours 43 mins
The MacBook Pro M1 max took 2 hours 46 mins.
List of tools tested https://macinchem.co.uk/software-reviews/cheminformatics-and-compchem-on-apple-silicon/
Last update 4 July 2023