A recent paper published in Nature caught my eye, Accurate predictions on small data with a tabular foundation model by Hollmann et al.,
Here we present the Tabular Prior-data Fitted Network (TabPFN), a tabular foundation model that outperforms all previous methods on datasets with up to 10,000 samples by a wide margin, using substantially less training time.
This foundation model was trained on around 130,000,000 synthetically generated datasets that mimic “real world” tabular data. These datasets sampled dataset size and number of features, both classification and regression tasks, and Gaussian noise was added to mimic real-world complexities.
They report TabPFN excels in handling small- to medium-sized datasets with up to 10,000 samples and 500 features, this is actually ideal for many projects. Indeed, whilst there is a huge amount of interest in very, very large global models in many cases a smaller local model performs as well or better DOI.
There is a very nice exploration of TabPFN in a cheminformatics setting here TabPFN for chemical datasets that uses the Therapeutic data commons (TDC) and RDKit descriptors. They also provide the python script used. This was used with minor modifications to allow selection of gpu or cpu, and selection of particular datasets. some datasets failed on first pass and the input data needed to be cleaned.
To install
|
1 2 3 4 |
git clone https://github.com/jonswain/tabpfn-tdc.git cd tabpfn-tdc conda env create -f environment.yml conda activate tabpfn-tdc |
This creates a folder called tabpfn-tdc that contains all the data and submission python script that can be invoked with.
|
1 |
python submission.py | 2>&1 | tee -a log.tx |
The python script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 |
from time import time import pandas as pd import torch from rdkit import Chem from rdkit.Chem import Descriptors from tabpfn import TabPFNClassifier, TabPFNRegressor from tdc.benchmark_group import admet_group from tqdm import tqdm def calculateDescriptors(mol: Chem.Mol, missingVal: float | None = 0.0) -> dict: """Calculate the full list of descriptors for a molecule.""" res = {} for nm, fn in Descriptors._descList: try: val = fn(mol) except: val = missingVal res[nm] = val return res def createDescriptorDataFrame( smiles: list[str], max_value: float = torch.finfo(torch.half).max, ) -> pd.DataFrame: """Create a DataFrame of descriptors for a list of SMILES strings.""" mols = [Chem.MolFromSmiles(smi) for smi in smiles] descs = [calculateDescriptors(mol) for mol in mols] return pd.DataFrame(descs).clip(-max_value, max_value) def getDeviceType() -> str: """Get the device type to use for training and inference.""" if torch.cuda.is_available(): return "cuda" elif torch.backends.mps.is_available(): #comment out to use cpu return "mps" #comment out to use cpu else: return "cpu" def main(): """Train and evaluate the model on the benchmark datasets.""" # Get the device type to use for training and inference device_type = getDeviceType() print(f"Training and inference completed using: {device_type}") # Load the benchmark datasets group = admet_group(path="data/") predictions_list = [{}, {}, {}, {}, {}] for dataset_name in group.dataset_names: print(f"Dataset: {dataset_name}") start = time() benchmark = group.get(dataset_name) train_val, test = benchmark["train_val"], benchmark["test"] # Edit to select particular groups of datasets if len(train_val) > 1800: print(f"Skipping {dataset_name} due to size") continue if dataset_name != "lipophilicity_astrazeneca": print(f"Skipping {dataset_name} wrong name") continue X_train = createDescriptorDataFrame(train_val["Drug"]) y_train = train_val["Y"] X_test = createDescriptorDataFrame(test["Drug"]) y_test = test["Y"] print(f"Train: {X_train.shape}, {y_train.shape}") print(f"Test: {X_test.shape}, {y_test.shape}") for seed in tqdm([1, 2, 3, 4, 5]): params = { "random_state": seed, "n_jobs": -1, "n_estimators": 4, "device": device_type, } # TODO: Work out how to use memory saving with MPS if device_type == "mps": params["memory_saving_mode"] = True params["ignore_pretraining_limits"] = True # TabPFN has a maximum number of samples it can handle if len(X_train) > 10000: X_train = X_train.sample(10000) y_train = y_train.loc[X_train.index] if y_test.nunique() == 2: model = TabPFNClassifier(**params) model.fit(X_train, y_train) y_pred = model.predict_proba(X_test)[:, 1] else: model = TabPFNRegressor(**params) model.fit(X_train, y_train) y_pred = model.predict(X_test) predictions_list[seed - 1][dataset_name] = y_pred ave, std = group.evaluate_many(predictions_list)[dataset_name] print(f"Performance: {ave:.3f} +/- {std:.3f}") end = time() print(f"Average time taken: {(end - start)/5:.2f} s") performance = group.evaluate_many( predictions_list, save_file_name="submission.txt" ) print(performance) if __name__ == "__main__": main() |
Results
The results are shown in the table below, the TabFPN performance matches that shown previously.
The first time I ran it a number of the datasets failed and on closer inspection a number of the records needed to be removed or edited. For example
“Butanal, reaction products with aniline”,CCCC=O.Nc1ccccc1,-4.5021013295
dialuminium(3+) ion dimolybdenum nonaoxidandiide,[Al+3].[Al+3].[Mo].[Mo].[O-2].[O-2].[O-2].[O-2].[O-2].[O-2].[O-2].[O-2].[O-2],-4.2291529922
The performance using gpu and cpu was identical and in many cases matched or exceeded the methods described in the TDC leaderboard.
| M2 Mac Studio Ultra | ||||||||
| Dataset | Size | Task | Metric | TabFPN performance | Current TDC best performance | TabPFN TDC leaderboard rank | Ave Time (mins) GPU | Ave Time (mins) CPU |
| Caco2_Wang | 906 | Regression | MAE | 0.282 ± 0.005 | 0.276 ± 0.005 | 2nd | 2.1 | 1.3 |
| HIA_Hou | 578 | Classification | AUROC | 0.987 ± 0.001 | 0.990 ± 0.002 | 5th | 1.2 | 0.5 |
| Pgp_Broccatelli | 1218 | Classification | AUROC | 0.937± 0.004 | 0.938 ± 0.002 | 2nd | 2.9 | 2.2 |
| Bioavailability_Ma | 640 | Classification | AUROC | 0.732 ± 0.016 | 0.753 ± 0.000 | 5th | 1.4 | 0.73 |
| Bbb_Martins | 2030 | Classification | AUROC | 0.918 ± 0.003 | 0.920 ± 0.006 | 2nd | 12.7 | 5.75 |
| Vdss_Lombardo | 1130 | Regression | Spearman | 0.693 ± 0.004 | 0.713 ± 0.007 | 3rd | 2.9 | 2.2 |
| Cyp2D6_Substrate_Carbonmangels | 667 | Classification | AUPRC | 0.717 ± 0.009 | 0.736 | 6th | 4 | 0.8 |
| Cyp3A4_Substrate_Carbonmangels | 670 | Classification | AUROC | 0.641 ± 0.004 | 0.667 ± 0.019 | 7th | 4.1 | 0.9 |
| Cyp2C9_Substrate_Carbonmangels | 669 | Classification | AUPRC | 0.400 ± 0.013 | 0.441 ± 0.033 | 10th | 4.1 | 0.85 |
| Half_Life_Obach | 667 | Regression | Spearman | 0.546 ± 0.013 | 0.576 ± 0.025 | 6th | 4.1 | 0.8 |
| Clearance_Microsome_Az | 1102 | Regression | Spearman | 0.632 ± 0.006 | 0.630 ± 0.010 | 1st | 6.9 | 1.9 |
| Clearance_Hepatocyte_Az | 1213 | Regression | Spearman | 0.396 ± 0.004 | 0.536 ± 0.02 | >10th | 7.6 | 2.4 |
| Herg | 655 | Classification | AUROC | 0.850 ± 0.002 | 0.880 ± 0.002 | 6th | 4 | 0.8 |
| Dili | 475 | Classification | AUROC | 0.910 ± 0.005 | 0.925 ± 0.005 | 6th | 3 | 0.5 |
| Larger Datasets | ||||||||
| lipophilicity_astrazeneca | 4200 | Regression | MAE | 0.506 ± 0.005 | 0.46 ± 0.006 | 5th | 24 | 22.9 |
| ppbr_az | 2790 | Regression | MAE | 7.075 +/- 0.035 | 7.505 ± 0.073 | 1st | 17.6 | 10.7 |
| AMES | 7255 | Binary | AUROC | 0.845 ± 0.002 | 0.871 ± 0.002 | 8th | 64 | 63 |
| solubility_aqsoldb | 9982 | Regression | MAE | 0.756 +/- 0.003 | 0.725 ± 0.011 | 2nd | 117 | 115 |
| LD50_Zhu | 7385 | Regression | MAE | 0.603 +/- 0.004 | 0.541 ± 0.015 | 4th | 69 | 69 |
| M1 MacBook Pro Max | ||||||||
| HIA_Hou | 578 | Classification | AuROC | 0.986 ± 0.001 | 0.990 ± 0.002 | 5th | 3.5 | 0.76 |
| Clearance_Microsome_Az | 1213 | Regression | MAE | 0.630 ± 0.006 | 0.630 ± 0.005 | 1st | 6.7 | 2.3 |
Rather unexpectedly the time taken using cpu for the initial tests was much less than using gpu (mps). I did wonder if I’d got the data switched, but repeating and checking cpu usage as shown below.
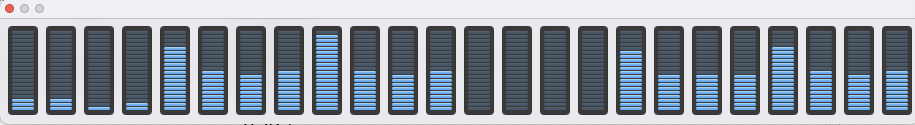
In contrast when using mps the gpu usage shot up
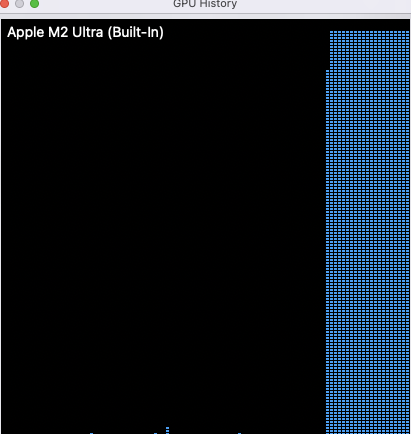
However for the larger data sets the times became comparable and around 7500 records the time taken is the same. Whilst inference is not as demanding as building the model it is not clear why cpu is better for the smaller data sets than gpu. As data sets get larger it might be expected that parallelisation on gpu might be advantageous, but I’ll investigate this in a subsequent post.
I also had a look at the performance on a M1 MacBook Pro Max, it was a little slower but still useful.
I’ve looked at a number of other cheminformatics tools on Apple Silicon here https://macinchem.org/cheminformatics-and-compchem-on-apple-silicon/