In a previous tutorial we made use of the Virtual Computational Chemistry Laboratory web service to calculate aLogP and LogS, both these results were returned in a simple text format. More recently there has been an increased use of JSON format for data exchange.
JSON, or JavaScript Object Notation, is a text-based open standard designed for easy human-readable data interchange. It is derived from the JavaScript scripting language for representing simple data structures and associative arrays, called objects. Despite its relationship to JavaScript, it is language-independent, with parsers available for many languages including including C, C++, C#, Java, JavaScript, Perl, Python.
JSON is built on two structures
- A collection of name/value pairs. In various languages, this is realised as an object, record, struct, dictionary, hash table, keyed list, or associative array.
- An ordered list of values. In most languages, this is realised as an array, vector, list, or sequence.
JSON can be used to provide the results of a calculation via a web service or it could be used to provide the results of a database search, indeed the popular database mongoDB uses JSON-style document storage. It can be regarded as less verbose alternative to XML.
Unfortunately the current version of Jython (2.5.x) does not support JSON however there is A fast JSON opensource codec for jython 2.5, written in java called jyson.
Installing jyson
You can download jyson from here, I’m not sure where the best place to install the jyson-1.0.2.jar file is, but I put it in “/Users/username/vortex/scripts/lib/jyson-1.0.2.jar. You need to make a note of where you put it because you need the full path to include in the script.
Molinspiration web services
Molinspiration provide a number of cheminformatics tools but also provide a RESTful web service these web services can be used to calculate a range of molecular properties and bioactivity predictions.
The output from both web services is available either as a JSON string or plain text, the web service can be accessed by submitting a URL in the following format, noting that the SMILES string needs to be encoded
|
1 2 |
http://www.molinspiration.com/cgi-bin/properties?smiles=URLEncodedSmiles&out=json&type=pr |
If bioactivity is required simply change to &type=ba
The result is returned in JSON format
|
1 2 |
{"status":"OK","smiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","smsmiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","natoms":13,"MW":244.044,"PSA":62.895,"logP":2.532,"nrotb":2,"nON":4,"nOHNH":0,"volume":160.808,"nviolations":0} |
Where the property name and value appear as colon separated pairs.
Note:-
The limit for a number of calculations for an IP address is 1000 per month to assure good availability of services for all users.
Explanation
Since we are going to be using an external java module we need to tell the script where the jar file is and then import it. You will need to modify the path to the jar file.
|
1 2 3 4 |
# This path to the jyson-1.0.2.jar will need to be modified sys.path.append("/Users/*your username*/vortex/scripts/lib/jyson-1.0.2.jar") from com.xhaus.jyson import JysonCodec as json |
Since this script access an external web service it is probably not a good idea to send your proprietary structures to this service, so the next part of the script reminds the user of this, if the user presses OK then the script runs.
The next part of the script sets up all the columns and sets the column type to double.
We then need to get the SMILES string for each structure as we loop through the table.
|
1 2 |
smiles = vortex.getMolProperty(mfm.getMolFileAtRow(r), 'SMILES') |
The next task is to generate the URL containing the SMILES string, however since SMILES often contain characters outside the ASCII set, the SMILES string has to be converted into a valid ASCII format. The URL encoding replaces unsafe ASCII characters with a “%” followed by two hexadecimal digits.
We then submit the data to the web service and capture the output in JSON format (shown below)
|
1 2 |
{"status":"OK","smiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","smsmiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","natoms":13,"MW":244.044,"PSA":62.895,"logP":2.532,"nrotb":2,"nON":4,"nOHNH":0,"volume":160.808,"nviolations":0} |
One useful feature to aid error checking is to include the print() command, this prints the results to the Jython console (you can open the Console from the Scripts menu).
|
1 2 3 |
j = json.loads(myreturn) print(j) |
The print command results in this appearing in the console.
|
1 2 |
{'PSA': 17.071, 'natoms': 13, 'nviolations': 0, 'logP': 2.731, 'status': u'OK', 'volume': 150.885, 'smsmiles': u'CC(=O)c1ccc(C(F)(F)F)cc1', 'MW': 188.148, 'nOHNH': 0, 'nrotb': 2, 'smiles': u'CC(=O)c1ccc(C(F)(F)F)cc1', 'nON': 1} |
It is important to note that the order of objects is not the same as returned by the web service and to access each of them when need to use the object name.
e,g.
|
1 2 |
logp = j['logP'] |
One last minor issue is Vortex uses setValueFromString because it is most reusable as you don’t then have to code for data types, this does mean however that we need to convert the values to strings before importing them into the table.
|
1 2 |
collogp.setValueFromString(r, str(j['logP'])) |
Finally we update the table.
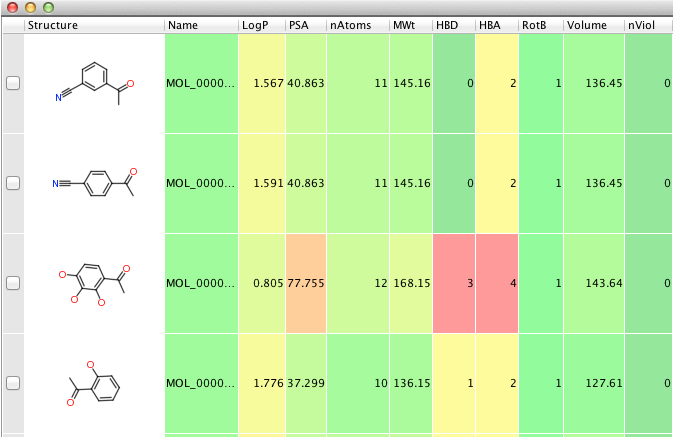
The Molinspiration properties Vortex Script
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 |
# This uses the Molinspiration web service (http://www.molinspiration.com) # http://www.molinspiration.com/cgi-bin/properties?smiles=c1cnccc1&out=json&type=pr import sys import com.dotmatics.vortex.util.Util as Util # This path to the jyson-1.0.2.jar will need to be modified sys.path.append("/Users/*your username*/vortex/scripts/lib/jyson-1.0.2.jar") from com.xhaus.jyson import JysonCodec as json if Util.getPlatform() == Util.PlatformIsWindows: sys.path.append(vortex.getVortexFolder() + '\\modules\\jythonconsole') sys.path.append(vortex.getVortexFolder() + '\\modules\\jythonlib') else: sys.path.append(vortex.getVortexFolder() + '/modules/jythonconsole') sys.path.append(vortex.getVortexFolder() + '/modules/jythonlib') import urllib2 import urllib content = javax.swing.JPanel() label = javax.swing.JLabel("<html><b>Calculate properties using the http://www.molinspiration.com web service</b><p><br>If you don't want to post your data to the internet press Cancel.</html>") layout.fill(content, label, 2, 2) ret = vortex.showInDialog(content, "Molinspiration Web Service") if ret == vortex.OK: collogp = vtable.findColumnWithName("LogP", 1, 1) colpsa = vtable.findColumnWithName("PSA", 1, 1) colnatoms = vtable.findColumnWithName("nAtoms", 1, 1) colMWt = vtable.findColumnWithName("MWt", 1, 1) colnOHNH = vtable.findColumnWithName("HBD", 1, 1) colnON = vtable.findColumnWithName("HBA", 1, 1) colnrotb = vtable.findColumnWithName("RotB", 1, 1) colvol = vtable.findColumnWithName("Volume", 1, 1) colnviolations = vtable.findColumnWithName("nViol", 1, 1) mfm = vtable.getMolFileManager() rows = vtable.getRealRowCount() for r in range(0, rows): smiles = vortex.getMolProperty(mfm.getMolFileAtRow(r), 'SMILES') encodedsmiles = urllib.urlencode({'':smiles}) mystr = "http://www.molinspiration.com/cgi-bin/properties?smiles" + encodedsmiles + "&out=json&type=pr" try: myreturn = urllib2.urlopen(mystr).read() if (len(myreturn) > 0): #parse data #['{"status":"OK","smiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","smsmiles":"CC(=O)c1ccc(Br)c(N(=O)=O)c1","natoms":13,"MW":244.044,"PSA":62.895,"logP":2.532,"nrotb":2,"nON":4,"nOHNH":0,"volume":160.808,"nviolations":0}\r\n', '\n'] j = json.loads(myreturn) #print(j) collogp.setValueFromString(r, str(j['logP'])) colpsa.setValueFromString(r, str(j['PSA'])) colnatoms.setValueFromString(r, str(j['natoms'])) colMWt.setValueFromString(r, str(j['MW'])) colnOHNH.setValueFromString(r, str(j['nOHNH'])) colnON.setValueFromString(r, str(j['nON'])) colnrotb.setValueFromString(r, str(j['nrotb'])) colvol.setValueFromString(r, str(j['volume'])) colnviolations.setValueFromString(r, str(j['nviolations'])) except: pass vtable.fireTableStructureChanged() |
The vortex script can be downloaded from here
The Molinspiration predicted biological activity Vortex Script
This is a modification of the above script and accesses the predicted biological properties.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 |
# This uses the Molinspiration web service (http://www.molinspiration.com) # http://www.molinspiration.com/cgi-bin/properties?smiles=c1cnccc1&out=json&type=ba import sys import com.dotmatics.vortex.util.Util as Util # This path to the jyson-1.0.2.jar will need to be modified sys.path.append("/Users/*your username*/vortex/scripts/lib/jyson-1.0.2.jar") from com.xhaus.jyson import JysonCodec as json if Util.getPlatform() == Util.PlatformIsWindows: sys.path.append(vortex.getVortexFolder() + '\\modules\\jythonconsole') sys.path.append(vortex.getVortexFolder() + '\\modules\\jythonlib') else: sys.path.append(vortex.getVortexFolder() + '/modules/jythonconsole') sys.path.append(vortex.getVortexFolder() + '/modules/jythonlib') import urllib2 import urllib content = javax.swing.JPanel() label = javax.swing.JLabel("<html><b>Calculate properties using the http://www.molinspiration.com web service</b><p><br>If you don't want to post your data to the internet press Cancel.</html>") layout.fill(content, label, 2, 2) ret = vortex.showInDialog(content, "Molinspiration Web Service") if ret == vortex.OK: colgpcrscore = vtable.findColumnWithName("gpcrscore", 1, 1) colicscore = vtable.findColumnWithName("icscore", 1, 1) colkiscore = vtable.findColumnWithName("kiscore", 1, 1) colnrscore = vtable.findColumnWithName("nrscore", 1, 1) colprscore = vtable.findColumnWithName("prscore", 1, 1) colezscore = vtable.findColumnWithName("ezscore", 1, 1) mfm = vtable.getMolFileManager() rows = vtable.getRealRowCount() for r in range(0, rows): smiles = vortex.getMolProperty(mfm.getMolFileAtRow(r), 'SMILES') encodedsmiles = urllib.urlencode({'':smiles}) mystr = "http://www.molinspiration.com/cgi-bin/properties?smiles" + encodedsmiles + "&out=json&type=ba" try: myreturn = urllib2.urlopen(mystr).read() if (len(myreturn) > 0): #parse data #{"status":"OK","smiles":"c1ccncc1","smsmiles":"c1ccncc1","gpcrscore":-3.7367,"icscore":-3.4464,"kiscore":-3.5241,"nrscore":-4.1832,"prscore":-3.8923,"ezscore":-3.5620} j = json.loads(myreturn) print(j) colgpcrscore.setValueFromString(r, str(j['gpcrscore'])) colicscore.setValueFromString(r, str(j['icscore'])) colkiscore.setValueFromString(r, str(j['kiscore'])) colnrscore.setValueFromString(r, str(j['nrscore'])) colprscore.setValueFromString(r, str(j['prscore'])) colezscore.setValueFromString(r, str(j['ezscore'])) except: pass vtable.fireTableStructureChanged() |
The vortex script can be downloaded from here
Last updated 21 February 2013